Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants
- PMID: 16569856
- PMCID: PMC1426987
- DOI: 10.1128/AAC.50.4.1384-1392.2006
Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants
Abstract
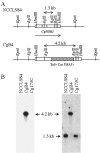
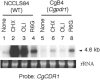
Candida glabrata, a yeast with intrinsically low susceptibility to azoles, frequently develops increased azole resistance during prolonged treatment. Transposon mutagenesis revealed that disruption of CgPDR1 resulted in an 8- to 16-fold increase in fluconazole susceptibility of C. glabrata. CgPDR1 is a homolog of Saccharomyces cerevisiae PDR1, which encodes a transcriptional regulator of multidrug transporters. Northern blot analyses indicated that CgPDR1 regulated both constitutive and drug-induced expression of CgCDR1, a multidrug transporter gene. In agreement with the Northern analysis, the Cgpdr1 mutant had increased rhodamine accumulation, in contrast to the decreased accumulation in the CgPDR1-overexpressing strain. Northern analyses also indicated the importance of CgPDR1 in fluconazole resistance arising during therapy. Two clinically resistant isolates had higher expression of CgPDR1 and CgCDR1 compared to their paired susceptible isolates. Integrative transformation of CgPDR1 from the two resistant isolates converted the Cgpdr1 mutant into azole-resistant strains with upregulated CgPDR1 expression. Two different amino acid substitutions, W297S in one isolate and F575L in the other, accounted for the upregulated CgPDR1 expression and the resistance. Finally, CgPDR1 was shown to be required for the azole resistance due to mitochondrial deficiency. Thus, CgPDR1 encodes a transcriptional regulator of a pleiotropic drug resistance network and contributes to the azole resistance of clinical isolates and petite mutants.
Figures






Similar articles
-
Microarray and molecular analyses of the azole resistance mechanism in Candida glabrata oropharyngeal isolates.Antimicrob Agents Chemother. 2010 Aug;54(8):3308-17. doi: 10.1128/AAC.00535-10. Epub 2010 Jun 14. Antimicrob Agents Chemother. 2010. PMID: 20547810 Free PMC article.
-
Missense mutation in CgPDR1 regulator associated with azole-resistant Candida glabrata recovered from Thai oral candidiasis patients.J Glob Antimicrob Resist. 2019 Jun;17:221-226. doi: 10.1016/j.jgar.2019.01.006. Epub 2019 Jan 15. J Glob Antimicrob Resist. 2019. PMID: 30658200
-
Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor.Antimicrob Agents Chemother. 2004 Oct;48(10):3773-81. doi: 10.1128/AAC.48.10.3773-3781.2004. Antimicrob Agents Chemother. 2004. PMID: 15388433 Free PMC article.
-
Multiple interfaces control activity of the Candida glabrata Pdr1 transcription factor mediating azole drug resistance.Curr Genet. 2019 Feb;65(1):103-108. doi: 10.1007/s00294-018-0870-4. Epub 2018 Jul 28. Curr Genet. 2019. PMID: 30056490 Free PMC article. Review.
-
Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment.Dan Med J. 2016 Oct;63(10):B5288. Dan Med J. 2016. PMID: 27697142 Review.
Cited by
-
Two Functionally Redundant FK506-Binding Proteins Regulate Multidrug Resistance Gene Expression and Govern Azole Antifungal Resistance.Antimicrob Agents Chemother. 2021 May 18;65(6):e02415-20. doi: 10.1128/AAC.02415-20. Print 2021 May 18. Antimicrob Agents Chemother. 2021. PMID: 33722894 Free PMC article.
-
Modulators of Candida albicans Membrane Drug Transporters: A Lucrative Portfolio for the Development of Effective Antifungals.Mol Biotechnol. 2024 May;66(5):960-974. doi: 10.1007/s12033-023-01017-1. Epub 2024 Jan 11. Mol Biotechnol. 2024. PMID: 38206530 Review.
-
Candida infections, causes, targets, and resistance mechanisms: traditional and alternative antifungal agents.Biomed Res Int. 2013;2013:204237. doi: 10.1155/2013/204237. Epub 2013 Jun 26. Biomed Res Int. 2013. PMID: 23878798 Free PMC article. Review.
-
Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond.Chem Rev. 2021 Mar 24;121(6):3390-3411. doi: 10.1021/acs.chemrev.0c00199. Epub 2020 May 22. Chem Rev. 2021. PMID: 32441527 Free PMC article. Review.
-
Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon.Eukaryot Cell. 2011 Mar;10(3):373-83. doi: 10.1128/EC.00073-10. Epub 2010 Dec 30. Eukaryot Cell. 2011. PMID: 21193550 Free PMC article.
References
-
- Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262:16871-16879. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

