Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA
- PMID: 16569869
- PMCID: PMC1426975
- DOI: 10.1128/AAC.50.4.1489-1496.2006
Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA
Abstract
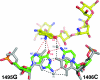
Aminoglycoside antibiotics that bind to the aminoacyl-tRNA site (A site) of the ribosome are composed of a common neamine core in which a glycopyranosyl ring is attached to position 4 of a 2-deoxystreptamine moiety. The core is further substituted by one (ribostamycin), two (neomycin and paromomycin), or three (lividomycin A) additional sugars attached to position 5 of the 2-deoxystreptamine. To study the role of rings III, IV, and V in aminoglycoside binding, we used isogenic Mycobacterium smegmatis DeltarrnB mutants carrying homogeneous populations of mutant ribosomes with alterations in the 16S rRNA A site. MICs were determined to investigate drug-ribosome interactions, and the results were compared with that of the previously published crystal structure of paromomycin bound to the ribosomal A site. Our analysis demonstrates that the stacking interaction between ring I and G1491 is largely sequence independent, that rings III and IV each increase the strength of drug binding to the ribosome, that ring IV of the 6'-NH3+ aminoglycosides compensates for loss of interactions between ring II and U1495 and between ring III and G1491, that the aminoglycosides rely on pseudo-base pairing between ring I and A1408 for binding independently of the number of sugar rings attached to the neamine core, that addition of ring V to the 6'-OH 4,5-aminoglycoside paromomycin does not alter the mode of binding, and that alteration of the U1406.U1495 wobble base pair to the Watson-Crick interaction pair 1406C-1495G yields ribosomal drug susceptibilities to 4,5-aminoglycosides comparable to those seen with the wild-type A site.
Figures




Similar articles
-
The molecular basis for A-site mutations conferring aminoglycoside resistance: relationship between ribosomal susceptibility and X-ray crystal structures.Chembiochem. 2003 Oct 6;4(10):1078-88. doi: 10.1002/cbic.200300657. Chembiochem. 2003. PMID: 14523926
-
Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6'OH and 6'NH3+ aminoglycosides.J Mol Biol. 2005 Feb 18;346(2):467-75. doi: 10.1016/j.jmb.2004.11.073. Epub 2004 Dec 21. J Mol Biol. 2005. PMID: 15670597
-
Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA.J Mol Biol. 1998 Mar 27;277(2):347-62. doi: 10.1006/jmbi.1997.1552. J Mol Biol. 1998. PMID: 9514735
-
The ribosomal A-site as an inspiration for the design of RNA binders.Biochimie. 2006 Aug;88(8):1045-51. doi: 10.1016/j.biochi.2006.03.005. Epub 2006 Mar 27. Biochimie. 2006. PMID: 16581175 Review.
-
A genetic model to investigate drug-target interactions at the ribosomal decoding site.Biochimie. 2006 Aug;88(8):1033-43. doi: 10.1016/j.biochi.2006.04.008. Epub 2006 Apr 27. Biochimie. 2006. PMID: 16690195 Review.
Cited by
-
Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity.RNA. 2008 Jan;14(1):148-57. doi: 10.1261/rna.805208. Epub 2007 Nov 14. RNA. 2008. PMID: 18003936 Free PMC article.
-
Roles of specific aminoglycoside-ribosome interactions in the inhibition of translation.RNA. 2019 Feb;25(2):247-254. doi: 10.1261/rna.068460.118. Epub 2018 Nov 9. RNA. 2019. PMID: 30413565 Free PMC article.
-
Understanding the origins of bacterial resistance to aminoglycosides through molecular dynamics mutational study of the ribosomal A-site.PLoS Comput Biol. 2011 Jul;7(7):e1002099. doi: 10.1371/journal.pcbi.1002099. Epub 2011 Jul 21. PLoS Comput Biol. 2011. PMID: 21814503 Free PMC article.
-
Hibernating ribosomes as drug targets?Front Microbiol. 2024 Jul 29;15:1436579. doi: 10.3389/fmicb.2024.1436579. eCollection 2024. Front Microbiol. 2024. PMID: 39135874 Free PMC article. Review.
-
Genetic reconstruction of protozoan rRNA decoding sites provides a rationale for paromomycin activity against Leishmania and Trypanosoma.PLoS Negl Trop Dis. 2011 May;5(5):e1161. doi: 10.1371/journal.pntd.0001161. Epub 2011 May 24. PLoS Negl Trop Dis. 2011. PMID: 21629725 Free PMC article.
References
-
- Auffinger, P., and E. Westhof. 2001. Water and ion binding around r(UpA)12 and d(TpA)12 oligomers—comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol. 305:1057-1072. - PubMed
-
- Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. - PubMed
-
- Dahlberg, A. E. 1989. The functional role of ribosomal RNA in protein synthesis. Cell 57:525-529. - PubMed
-
- Davies, J., L. Gorini, and B. D. Davis. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93-106. - PubMed
-
- De Stasio, E. A., and A. E. Dahlberg. 1990. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J. Mol. Biol. 212:127-133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

