Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission
- PMID: 16571797
- PMCID: PMC1440464
- DOI: 10.1128/JVI.80.8.3801-3810.2006
Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission
Abstract
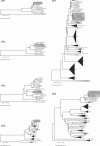
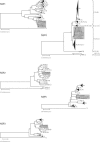
The Belgian rotavirus strain B4106, isolated from a child with gastroenteritis, was previously found to have VP7 (G3), VP4 (P[14]), and NSP4 (A genotype) genes closely related to those of lapine rotaviruses, suggesting a possible lapine origin or natural reassortment of strain B4106. To investigate the origin of this unusual strain, the gene sequences encoding VP1, VP2, VP3, VP6, NSP1, NSP2, NSP3, and NSP5/6 were also determined. To allow comparison to a lapine strain, the 11 double-stranded RNA segments of a European G3P[14] rabbit rotavirus strain 30/96 were also determined. The complete genome similarity between strains B4106 and 30/96 was 93.4% at the nucleotide level and 96.9% at the amino acid level. All 11 genome segments of strain B4106 were closely related to those of lapine rotaviruses and clustered with the lapine strains in phylogenetic analyses. In addition, sequence analyses of the NSP5 gene of strain B4106 revealed that the altered electrophoretic mobility of NSP5, resulting in a super-short pattern, was due to a gene rearrangement (head-to-tail partial duplication, combined with two short insertions and a deletion). Altogether, these findings confirm that a rotavirus strain with an entirely lapine genome complement was able to infect and cause severe disease in a human child.
Figures

References
-
- Ahmed, K., T. Nakagomi, and O. Nakagomi. 2005. Isolation and molecular characterization of a naturally occurring non-structural protein 5 (NSP5) gene reassortant of group A rotavirus of serotype G2P[4] with a long RNA pattern. J. Med. Virol. 77:323-330. - PubMed
-
- Aijaz, S., K. Gowda, H. V. Jagannath, R. R. Reddy, P. P. Maiya, R. L. Ward, H. B. Greenberg, M. Raju, A. Babu, and C. D. Rao. 1996. Epidemiology of symptomatic human rotaviruses in Bangalore and Mysore, India, from 1988 to 1994 as determined by electropherotype, subgroup, and serotype analysis. Arch. Virol. 141:715-726. - PubMed
-
- Alfieri, A. A., J. P. Leite, O. Nakagomi, E. Kaga, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Characterization of human rotavirus genotype P[8]G5 from Brazil by probe-hybridization and sequence. Arch. Virol. 141:2353-2364. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

