Characterization of functional domains of equine infectious anemia virus Rev suggests a bipartite RNA-binding domain
- PMID: 16571801
- PMCID: PMC1440447
- DOI: 10.1128/JVI.80.8.3844-3852.2006
Characterization of functional domains of equine infectious anemia virus Rev suggests a bipartite RNA-binding domain
Abstract
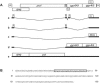
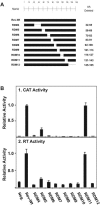
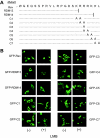
Equine infectious anemia virus (EIAV) Rev is an essential regulatory protein that facilitates expression of viral mRNAs encoding structural proteins and genomic RNA and regulates alternative splicing of the bicistronic tat/rev mRNA. EIAV Rev is characterized by a high rate of genetic variation in vivo, and changes in Rev genotype and phenotype have been shown to coincide with changes in clinical disease. To better understand how genetic variation alters Rev phenotype, we undertook deletion and mutational analyses to map functional domains and to identify specific motifs that are essential for EIAV Rev activity. All functional domains are contained within the second exon of EIAV Rev. The overall organization of domains within Rev exon 2 includes a nuclear export signal, a large central region required for RNA binding, a nonessential region, and a C-terminal region required for both nuclear localization and RNA binding. Subcellular localization of green fluorescent protein-Rev mutants indicated that basic residues within the KRRRK motif in the C-terminal region of Rev are necessary for targeting of Rev to the nucleus. Two separate regions of Rev were necessary for RNA binding: a central region encompassing residues 57 to 130 and a C-terminal region spanning residues 144 to 165. Within these regions were two distinct, short arginine-rich motifs essential for RNA binding, including an RRDRW motif in the central region and the KRRRK motif near the C terminus. These findings suggest that EIAV Rev utilizes a bipartite RNA-binding domain.
Figures

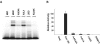
Similar articles
-
Computational modeling suggests dimerization of equine infectious anemia virus Rev is required for RNA binding.Retrovirology. 2014 Dec 23;11:115. doi: 10.1186/s12977-014-0115-7. Retrovirology. 2014. PMID: 25533001 Free PMC article.
-
Binding of equine infectious anemia virus rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs.Mol Cell Biol. 2000 May;20(10):3550-7. doi: 10.1128/MCB.20.10.3550-3557.2000. Mol Cell Biol. 2000. PMID: 10779344 Free PMC article.
-
Structural model of the Rev regulatory protein from equine infectious anemia virus.PLoS One. 2009;4(1):e4178. doi: 10.1371/journal.pone.0004178. Epub 2009 Jan 12. PLoS One. 2009. PMID: 19137065 Free PMC article.
-
Molecular and biological characterization of equine infectious anemia virus Rev.Curr HIV Res. 2010 Jan;8(1):87-93. doi: 10.2174/157016210790416424. Curr HIV Res. 2010. PMID: 20210784 Review.
-
Rev variation during persistent lentivirus infection.Viruses. 2011 Jan;3(1):1-11. doi: 10.3390/v3010001. Epub 2011 Jan 11. Viruses. 2011. PMID: 21994723 Free PMC article. Review.
Cited by
-
Computational modeling suggests dimerization of equine infectious anemia virus Rev is required for RNA binding.Retrovirology. 2014 Dec 23;11:115. doi: 10.1186/s12977-014-0115-7. Retrovirology. 2014. PMID: 25533001 Free PMC article.
-
Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism.J Virol. 2008 Dec;82(23):11889-901. doi: 10.1128/JVI.01537-08. Epub 2008 Sep 25. J Virol. 2008. PMID: 18818324 Free PMC article.
-
Characterization of Signal Sequences Determining the Nuclear/Nucleolar Import and Nuclear Export of the Caprine Arthritis-Encephalitis Virus Rev Protein.Viruses. 2020 Aug 17;12(8):900. doi: 10.3390/v12080900. Viruses. 2020. PMID: 32824614 Free PMC article.
-
The bovine immunodeficiency virus rev protein: identification of a novel lentiviral bipartite nuclear localization signal harboring an atypical spacer sequence.J Virol. 2009 Dec;83(24):12842-53. doi: 10.1128/JVI.01613-09. Epub 2009 Oct 14. J Virol. 2009. PMID: 19828621 Free PMC article.
-
Naturally arising point mutations in non-essential domains of equine infectious anemia virus Rev alter Rev-dependent nuclear-export activity.J Gen Virol. 2008 Apr;89(Pt 4):1043-1048. doi: 10.1099/vir.0.83195-0. J Gen Virol. 2008. PMID: 18343848 Free PMC article.
References
-
- Ali, S. A., and A. Steinkasserer. 1995. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18:746-750. - PubMed
-
- Beisel, C. E., J. F. Edwards, L. L. Dunn, and N. R. Rice. 1993. Analysis of multiple mRNAs from pathogenic equine infectious anemia virus (EIAV) in an acutely infected horse reveals a novel protein, ttm, derived from the carboxy terminus of the EIAV transmembrane protein. J. Virol. 67:832-842. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

