Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway
- PMID: 16571810
- PMCID: PMC1440455
- DOI: 10.1128/JVI.80.8.3935-3946.2006
Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway
Abstract
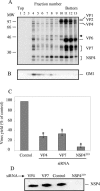
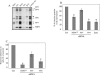
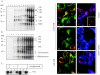
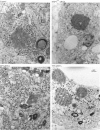
Studies of rotavirus morphogenesis, transport, and release have shown that although these viruses are released from the apical surface of polarized intestinal cells before cellular lysis, they do not follow the classic exocytic pathway. Furthermore, increasing evidence suggests that lipid rafts actively participate in the exit of rotavirus from the infected cell. In this study, we silenced the expression of VP4, VP7, and NSP4 by using small interfering RNAs (siRNAs) and evaluated the effect of shutting down the expression of these proteins on rotavirus-raft interactions. Silencing of VP4 and NSP4 reduced the association of rotavirus particles with rafts; in contrast, inhibition of VP7 synthesis slightly affected the migration of virions into rafts. We found that inhibition of rotavirus migration into lipid rafts, by either siRNAs or tunicamycin, also specifically blocked the targeting of VP4 to rafts, suggesting that the association of VP4 with rafts is mostly mediated by the formation of viral particles in the endoplasmic reticulum (ER). We showed that two populations of VP4 exist, one small population that is independently targeted to rafts and a second large pool of VP4 whose association with rafts is mediated by particle formation in the ER. We also present evidence to support the hypothesis that assembly of VP4 into mature virions takes place in the late stages of transit through the ER. Finally, we analyzed the progression of rotavirus proteins in the exocytic pathway and found that VP4 and virion-assembled VP7 colocalized with ERGIC-53, suggesting that rotavirus particles transit through the intermediate compartment between the ER and the Golgi complex.
Figures
Similar articles
-
Spike protein VP4 assembly with maturing rotavirus requires a postendoplasmic reticulum event in polarized caco-2 cells.J Virol. 2004 Oct;78(20):10987-94. doi: 10.1128/JVI.78.20.10987-10994.2004. J Virol. 2004. PMID: 15452219 Free PMC article.
-
Silencing the morphogenesis of rotavirus.J Virol. 2005 Jan;79(1):184-92. doi: 10.1128/JVI.79.1.184-192.2005. J Virol. 2005. PMID: 15596814 Free PMC article.
-
Actin-Dependent Nonlytic Rotavirus Exit and Infectious Virus Morphogenetic Pathway in Nonpolarized Cells.J Virol. 2018 Feb 26;92(6):e02076-17. doi: 10.1128/JVI.02076-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263265 Free PMC article.
-
Rotavirus assembly: an alternative model that utilizes an atypical trafficking pathway.Curr Top Microbiol Immunol. 2006;309:245-61. doi: 10.1007/3-540-30773-7_9. Curr Top Microbiol Immunol. 2006. PMID: 16909902 Free PMC article. Review.
-
A hypothesis about the mechanism of assembly of double-shelled rotavirus particles.Arch Virol Suppl. 1996;12:79-85. doi: 10.1007/978-3-7091-6553-9_9. Arch Virol Suppl. 1996. PMID: 9015104 Review.
Cited by
-
How viruses use the endoplasmic reticulum for entry, replication, and assembly.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a013250. doi: 10.1101/cshperspect.a013250. Cold Spring Harb Perspect Biol. 2013. PMID: 23284050 Free PMC article. Review.
-
Rotavirus infection activates the UPR but modulates its activity.Virol J. 2011 Jul 20;8:359. doi: 10.1186/1743-422X-8-359. Virol J. 2011. PMID: 21774819 Free PMC article.
-
Identification of a Small Molecule That Compromises the Structural Integrity of Viroplasms and Rotavirus Double-Layered Particles.J Virol. 2018 Jan 17;92(3):e01943-17. doi: 10.1128/JVI.01943-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142132 Free PMC article.
-
Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication.J Virol. 2010 Jul;84(13):6782-98. doi: 10.1128/JVI.01757-09. Epub 2010 Mar 24. J Virol. 2010. PMID: 20335253 Free PMC article.
-
Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome.Prog Lipid Res. 2010 Jan;49(1):1-26. doi: 10.1016/j.plipres.2009.07.003. Epub 2009 Jul 26. Prog Lipid Res. 2010. PMID: 19638285 Free PMC article. Review.
References
-
- Ben-Tekaya, H., K. Miura, R. Pepperkok, and H. P. Hauri. 2005. Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 118:357-367. - PubMed
-
- Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

