The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target
- PMID: 16571812
- PMCID: PMC1440456
- DOI: 10.1128/JVI.80.8.3957-3965.2006
The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target
Abstract
The emergence of influenza A viruses resistant to the two existing classes of antiviral drugs highlights the need for additional antiviral drugs, particularly considering the potential threat of a pandemic of H5N1 influenza A viruses. Here, we determine whether influenza A virus replication can be selectively inhibited by blocking the ability of its NS1A protein to inhibit the 3'-end processing of cellular pre-mRNAs, including beta interferon (IFN-beta) pre-mRNA. Pre-mRNA processing is inhibited via the binding of the NS1A protein to the cellular CPSF30 protein, and mutational inactivation of this NS1A binding site causes severe attenuation of the virus. We demonstrate that binding of CPSF30 is mediated by two of its zinc fingers, F2F3, and that the CPSF30/F2F3 binding site on the NS1A protein extends from amino acid 144 to amino acid 186. We generated MDCK cells that constitutively express epitope-tagged F2F3 in the nucleus, although at only approximately one-eighth the level of the NS1A protein produced during virus infection. Influenza A virus replication was inhibited in this cell line, whereas no inhibition was observed with influenza B virus, whose NS1B protein lacks a binding site for CPSF30. Influenza A virus, but not influenza B virus, induced increased production of IFN-beta mRNA in the F2F3-expressing cells. These results, which indicate that F2F3 inhibits influenza A virus replication by blocking the binding of endogenous CPSF30 to the NS1A protein, point to this NS1A binding site as a potential target for the development of antivirals directed against influenza A virus.
Figures
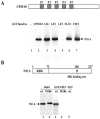
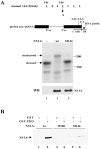



Similar articles
-
Structural basis for suppression of a host antiviral response by influenza A virus.Proc Natl Acad Sci U S A. 2008 Sep 2;105(35):13093-8. doi: 10.1073/pnas.0805213105. Epub 2008 Aug 25. Proc Natl Acad Sci U S A. 2008. PMID: 18725644 Free PMC article.
-
The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells.J Virol. 2007 Aug;81(15):8112-21. doi: 10.1128/JVI.00006-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522219 Free PMC article.
-
Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3' end processing of cellular pre-mRNAS.Virology. 2003 Mar 15;307(2):386-95. doi: 10.1016/s0042-6822(02)00127-7. Virology. 2003. PMID: 12667806
-
The influenza virus NS1 protein as a therapeutic target.Antiviral Res. 2013 Sep;99(3):409-16. doi: 10.1016/j.antiviral.2013.06.005. Epub 2013 Jun 21. Antiviral Res. 2013. PMID: 23796981 Free PMC article. Review.
-
Selective nuclear export of viral mRNAs in influenza-virus-infected cells.Trends Microbiol. 2000 Aug;8(8):376-83. doi: 10.1016/s0966-842x(00)01794-7. Trends Microbiol. 2000. PMID: 10920397 Review.
Cited by
-
Influenza virus A/Beijing/501/2009(H1N1) NS1 interacts with β-tubulin and induces disruption of the microtubule network and apoptosis on A549 cells.PLoS One. 2012;7(11):e48340. doi: 10.1371/journal.pone.0048340. Epub 2012 Nov 6. PLoS One. 2012. PMID: 23139776 Free PMC article.
-
The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2'-5' oligo (A) synthetase/RNase L pathway.Proc Natl Acad Sci U S A. 2006 May 2;103(18):7100-5. doi: 10.1073/pnas.0602184103. Epub 2006 Apr 20. Proc Natl Acad Sci U S A. 2006. PMID: 16627618 Free PMC article.
-
Structural basis for suppression of a host antiviral response by influenza A virus.Proc Natl Acad Sci U S A. 2008 Sep 2;105(35):13093-8. doi: 10.1073/pnas.0805213105. Epub 2008 Aug 25. Proc Natl Acad Sci U S A. 2008. PMID: 18725644 Free PMC article.
-
Natural Selection of H5N1 Avian Influenza A Viruses with Increased PA-X and NS1 Shutoff Activity.Viruses. 2021 Sep 3;13(9):1760. doi: 10.3390/v13091760. Viruses. 2021. PMID: 34578340 Free PMC article.
-
Influenza NS1 directly modulates Hedgehog signaling during infection.PLoS Pathog. 2017 Aug 24;13(8):e1006588. doi: 10.1371/journal.ppat.1006588. eCollection 2017 Aug. PLoS Pathog. 2017. PMID: 28837667 Free PMC article.
References
-
- Barabino, S. M., W. Hubner, A. Jenny, L. Minvielle-Sebastia, and W. Keller. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11:1703-1716. - PubMed
-
- Centers for Disease Control and Prevention. 28 September 2005. Key facts about influenza and the influenza vaccine. http://www.cdc.gov/flu/keyfacts.htm. [Online.]
-
- Cox, N. J., and K. Subbarao. 1999. Influenza. Lancet 354:1277-1282. - PubMed
-
- Ferguson, N. M., D. A. Cummings, S. Cauchemez, C. Fraser, S. Riley, A. Meeyai, S. Iamsirithaworn, and D. S. Burke. 2005. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437:209-214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

