Analysis of equid herpesvirus 1 strain variation reveals a point mutation of the DNA polymerase strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks
- PMID: 16571821
- PMCID: PMC1440451
- DOI: 10.1128/JVI.80.8.4047-4060.2006
Analysis of equid herpesvirus 1 strain variation reveals a point mutation of the DNA polymerase strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks
Abstract
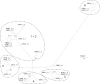
Equid herpesvirus 1 (EHV-1) can cause a wide spectrum of diseases ranging from inapparent respiratory infection to the induction of abortion and, in extreme cases, neurological disease resulting in paralysis and ultimately death. It has been suggested that distinct strains of EHV-1 that differ in pathogenic capacity circulate in the field. In order to investigate this hypothesis, it was necessary to identify genetic markers that allow subgroups of related strains to be identified. We have determined all of the genetic differences between a neuropathogenic strain (Ab4) and a nonneuropathogenic strain (V592) of EHV-1 and developed PCR/sequencing procedures enabling differentiation of EHV-1 strains circulating in the field. The results indicate the occurrence of several major genetic subgroups of EHV-1 among isolates recovered from outbreaks over the course of 30 years, consistent with the proposal that distinct strains of EHV-1 circulate in the field. Moreover, there is evidence that certain strain groups are geographically restricted, being recovered predominantly from outbreaks occurring in either North America or Europe. Significantly, variation of a single amino acid of the DNA polymerase is strongly associated with neurological versus nonneurological disease outbreaks. Strikingly, this variant amino acid occurs at a highly conserved position for herpesvirus DNA polymerases, suggesting an important functional role.
Figures

References
-
- Allen, G. P., J. H. Kydd, J. D. Slater, and K. C. Smith. 1999. Advances in understanding the pathogenesis, epidemiology and immunological control of equid herpes-1 (EHV-1) abortion, p. 129-146. In U. Wernery, J. F. Wade, J. A. Mumford, and O. R. Kaaden (ed.), Equine infectious diseases VIII. Proceedings of the Eighth International Conference on Equine Infectious Diseases. R&W Publications Limited, Newmarket, United Kingdom.
-
- Allen, G. P., M. R. Yeargan, L. W. Turtinen, and J. T. Bryans. 1985. A new field isolate of equine abortion virus (equine herpesvirus-1) among Kentucky horses. Am. J. Vet. Res. 46:138-140. - PubMed
-
- Allen, G. P., M. R. Yeargan, L. W. Turtinen, J. T. Bryans, and W. H. McCollum. 1983. Molecular epizootiologic studies of equine herpesvirus-1 infections by restriction endonuclease fingerprinting of viral DNA. Am. J. Vet. Res. 44:263-271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

