Expression of baculovirus late and very late genes depends on LEF-4, a component of the viral RNA polymerase whose guanyltransferase function is essential
- PMID: 16571832
- PMCID: PMC1440449
- DOI: 10.1128/JVI.80.8.4168-4173.2006
Expression of baculovirus late and very late genes depends on LEF-4, a component of the viral RNA polymerase whose guanyltransferase function is essential
Abstract
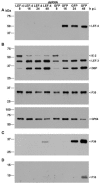
Baculovirus lef-4 encodes one subunit of the viral RNA polymerase. Here, we demonstrate the essential nature of LEF-4 by RNA interference and bacmid knockout technology. Silencing of LEF-4 in wild-type virus-infected cells suppressed expression of structural genes, while early expression was unaffected, demonstrating its essential role in late gene expression. After transfection of insect cells with lef-4 mutant bacmid, no viral progeny was produced, further defining its central role in infection. Cotransfection with wild-type lef-4 plasmid restored normal replication, but plasmid encoding a guanyltransferase-deficient version failed to rescue. These results emphasize the importance of the mRNA capping function of LEF-4.
Figures




Similar articles
-
Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication.J Virol. 2002 Mar;76(6):2770-9. doi: 10.1128/jvi.76.6.2770-2779.2002. J Virol. 2002. PMID: 11861844 Free PMC article.
-
Functional characterization of Bombyx mori nucleopolyhedrovirus mutant lacking late expression factor 9.Acta Virol. 2016;60(3):281-9. doi: 10.4149/av_2016_03_281. Acta Virol. 2016. PMID: 27640438
-
Characterization of late gene expression factor LEF-10 from Bombyx mori nucleopolyhedrovirus.Virus Res. 2013 Jul;175(1):45-51. doi: 10.1016/j.virusres.2013.03.022. Epub 2013 Apr 17. Virus Res. 2013. PMID: 23603137
-
An Autographa californica nucleopolyhedrovirus lef-2 mutant: consequences for DNA replication and very late gene expression.Virology. 1996 Mar 1;217(1):338-48. doi: 10.1006/viro.1996.0121. Virology. 1996. PMID: 8599220
-
Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV.Virology. 1997 Mar 31;230(1):35-47. doi: 10.1006/viro.1997.8457. Virology. 1997. PMID: 9126260
Cited by
-
Promoter motifs essential to the differential transcription of structural and non-structural genes of the white spot syndrome virus.Virus Genes. 2009 Oct;39(2):223-33. doi: 10.1007/s11262-009-0380-z. Virus Genes. 2009. PMID: 19554443
-
Use of bacterial artificial chromosomes in baculovirus research and recombinant protein expression: current trends and future perspectives.ISRN Microbiol. 2012 Sep 12;2012:628797. doi: 10.5402/2012/628797. Print 2012. ISRN Microbiol. 2012. PMID: 23762754 Free PMC article.
-
Systematic Analysis of 42 Autographa Californica Multiple Nucleopolyhedrovirus Genes Identifies An Additional Six Genes Involved in the Production of Infectious Budded Virus.Virol Sin. 2021 Aug;36(4):762-773. doi: 10.1007/s12250-021-00355-1. Epub 2021 Mar 8. Virol Sin. 2021. PMID: 33683665 Free PMC article.
-
The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects.J Virol. 2007 May;81(10):5395-406. doi: 10.1128/JVI.02781-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360757 Free PMC article.
-
Characterization of LEF4 ligand binding property and its role as part of baculoviral transcription machinery.Mol Cell Biochem. 2010 Jan;333(1-2):83-9. doi: 10.1007/s11010-009-0207-1. Epub 2009 Jul 25. Mol Cell Biochem. 2010. PMID: 19633819
References
-
- Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. - PubMed
-
- Carstens, E. B., H. Chan, H. Yu, G. V. Williams, and R. Casselman. 1994. Genetic analyses of temperature-sensitive mutations in baculovirus late expression factors. Virology 204:323-337. - PubMed
-
- Flores-Jasso, C. F., V. J. Valdes, A. Sampieri, V. Valadez-Graham, F. Recillas-Targa, and L. Vaca. 2004. Silencing structural and nonstructural genes in baculovirus by RNA interference. Virus Res. 102:75-84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

