The use of whole genome amplification to study chromosomal changes in prostate cancer: insights into genome-wide signature of preneoplasia associated with cancer progression
- PMID: 16573809
- PMCID: PMC1450280
- DOI: 10.1186/1471-2164-7-65
The use of whole genome amplification to study chromosomal changes in prostate cancer: insights into genome-wide signature of preneoplasia associated with cancer progression
Abstract
Background: Prostate cancer (CaP) is a disease with multifactorial etiology that includes both genetic and environmental components. The knowledge of the genetic basis of CaP has increased over the past years, mainly in the pathways that underlie tumourigenesis, progression and drug resistance. The vast majority of cases of CaP are adenocarcinomas that likely develop through a pre-malignant lesion and high-grade prostatic intraepithelial neoplasia (HPIN). Histologically, CaP is a heterogeneous disease consisting of multiple, discrete foci of invasive carcinoma and HPIN that are commonly interspersed with benign glands and stroma. This admixture with benign tissue can complicate genomic analyses in CaP. Specifically, when DNA is bulk-extracted the genetic information obtained represents an average for all of the cells within the sample.
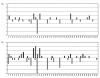
Results: To minimize this problem, we obtained DNA from individual foci of HPIN and CaP by laser capture microdissection (LCM). The small quantities of DNA thus obtained were then amplified by means of multiple-displacement amplification (MDA), for use in genomic DNA array comparative genomic hybridisation (gaCGH). Recurrent chromosome copy number abnormalities (CNAs) were observed in both HPIN and CaP. In HPIN, chromosomal imbalances involving chromosome 8 where common, whilst in CaP additional chromosomal changes involving chromosomes 6, 10, 13 and 16 where also frequently observed.
Conclusion: An overall increase in chromosomal changes was seen in CaP compared to HPIN, suggesting a universal breakdown in chromosomal stability. The accumulation of CNAs, which occurs during this process is non-random and may indicate chromosomal regions important in tumourigenesis. It is therefore likely that the alterations in copy number are part of a programmed cycle of events that promote tumour development, progression and survival. The combination of LCM, MDA and gaCGH is ideally suited for the identification of CNAs from small cell clusters and may assist in the discovery of potential genomic markers for early diagnosis, or identify the location of tumour suppressor genes (TSG) or oncogenes previously unreported in HPIN and CaP.
Figures
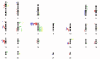
Similar articles
-
Use of whole genome amplification and comparative genomic hybridisation to detect chromosomal copy number alterations in cell line material and tumour tissue.Cytogenet Genome Res. 2004;105(1):18-24. doi: 10.1159/000078004. Cytogenet Genome Res. 2004. PMID: 15218253
-
Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma.Oncogene. 2003 Apr 3;22(13):1978-87. doi: 10.1038/sj.onc.1206227. Oncogene. 2003. PMID: 12673203
-
p53 Alteration and chromosomal instability in prostatic high-grade intraepithelial neoplasia and concurrent carcinoma: analysis by immunohistochemistry, interphase in situ hybridization, and sequencing of laser-captured microdissected specimens.Mod Pathol. 2001 Dec;14(12):1252-62. doi: 10.1038/modpathol.3880471. Mod Pathol. 2001. PMID: 11743048
-
Potential markers of aggressiveness in prostatic intraepithelial neoplasia detected by fluorescence in situ hybridization.Eur Urol. 1996;30(2):177-84. doi: 10.1159/000474167. Eur Urol. 1996. PMID: 8875198 Review.
-
Chromosome 6p amplification and cancer progression.J Clin Pathol. 2007 Jan;60(1):1-7. doi: 10.1136/jcp.2005.034389. Epub 2006 Jun 21. J Clin Pathol. 2007. PMID: 16790693 Free PMC article. Review.
Cited by
-
Recent advances and application of whole genome amplification in molecular diagnosis and medicine.MedComm (2020). 2022 Feb 3;3(1):e116. doi: 10.1002/mco2.116. eCollection 2022 Mar. MedComm (2020). 2022. PMID: 35281794 Free PMC article. Review.
-
Whole genome amplification and its impact on CGH array profiles.BMC Res Notes. 2008 Jul 29;1:56. doi: 10.1186/1756-0500-1-56. BMC Res Notes. 2008. PMID: 18710509 Free PMC article.
-
Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues.BMC Biotechnol. 2008 Feb 20;8:17. doi: 10.1186/1472-6750-8-17. BMC Biotechnol. 2008. PMID: 18284708 Free PMC article.
-
FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome.Br J Cancer. 2007 Sep 3;97(5):678-85. doi: 10.1038/sj.bjc.6603924. Epub 2007 Aug 14. Br J Cancer. 2007. PMID: 17700571 Free PMC article.
-
Estimating genomic instability mediated by Alu retroelements in breast cancer.Genet Mol Biol. 2009 Jan;32(1):25-31. doi: 10.1590/S1415-47572009005000018. Epub 2009 Jan 23. Genet Mol Biol. 2009. PMID: 21637642 Free PMC article.
References
-
- Al-Maghrabi J, Vorobyova L, Toi A, Chapman W, Zielenska M, Squire JA. Identification of numerical chromosomal changes detected by interphase fluorescence in situ hybridization in high-grade prostate intraepithelial neoplasia as a predictor of carcinoma. Arch Pathol Lab Med. 2002;126:165–169. - PubMed
-
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. - PubMed
-
- van Dekken H, Paris PL, Albertson DG, Alers JC, Andaya A, Kowbel D, van der Kwast TH, Pinkel D, Schroder FH, Vissers KJ, Wildhagen MF, Collins C. Evaluation of genetic patterns in different tumor areas of intermediate-grade prostatic adenocarcinomas by high-resolution genomic array analysis. Genes Chromosomes Cancer. 2004;39:249–256. doi: 10.1002/gcc.20001. - DOI - PubMed
-
- Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, Elkahloun A, Monni O, Chen Y, Kallioniemi A, Kallioniemi OP. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6:240–247. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

