A noncoding RNA is a potential marker of cell fate during mammary gland development
- PMID: 16574773
- PMCID: PMC1420634
- DOI: 10.1073/pnas.0600745103
A noncoding RNA is a potential marker of cell fate during mammary gland development
Abstract
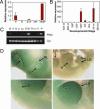
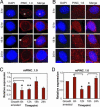
PINC is a large, alternatively spliced, developmentally regulated, noncoding RNA expressed in the regressed terminal ductal lobular unit-like structures of the parous mammary gland. Previous studies have shown that this population of cells possesses not only progenitor-like qualities (the ability to proliferate and repopulate a mammary gland) and the ability to survive developmentally programmed cell death but also the inhibition of carcinogen-induced proliferation. Here we report that PINC expression is temporally and spatially regulated in response to developmental stimuli in vivo and that PINC RNA is localized to distinct foci in either the nucleus or the cytoplasm in a cell-cycle-specific manner. Loss-of-function experiments suggest that PINC performs dual roles in cell survival and regulation of cell-cycle progression, suggesting that PINC may contribute to the developmentally mediated changes previously observed in the terminal ductal lobular unit-like structures of the parous gland. This is one of the first reports describing the functional properties of a large, developmentally regulated, mammalian, noncoding RNA.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation.PLoS Genet. 2012;8(7):e1002840. doi: 10.1371/journal.pgen.1002840. Epub 2012 Jul 26. PLoS Genet. 2012. PMID: 22911650 Free PMC article.
-
The long noncoding RNA Neat1 is required for mammary gland development and lactation.RNA. 2014 Dec;20(12):1844-9. doi: 10.1261/rna.047332.114. Epub 2014 Oct 14. RNA. 2014. PMID: 25316907 Free PMC article.
-
Expression and functional properties of transforming growth factor alpha and epidermal growth factor during mouse mammary gland ductal morphogenesis.Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):276-80. doi: 10.1073/pnas.88.1.276. Proc Natl Acad Sci U S A. 1991. PMID: 1986376 Free PMC article.
-
The concept of stem cell in the mammary gland and its implication in morphogenesis, cancer and prevention.Front Biosci. 2006 Jan 1;11:151-72. doi: 10.2741/1788. Front Biosci. 2006. PMID: 16146722 Review.
-
An atlas of mouse mammary gland development.J Mammary Gland Biol Neoplasia. 2000 Apr;5(2):227-41. doi: 10.1023/a:1026499523505. J Mammary Gland Biol Neoplasia. 2000. PMID: 11149575 Review.
Cited by
-
Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer.Oncotarget. 2016 Jun 28;7(26):40418-40436. doi: 10.18632/oncotarget.9622. Oncotarget. 2016. PMID: 27250026 Free PMC article.
-
On the classification of long non-coding RNAs.RNA Biol. 2013 Jun;10(6):925-33. doi: 10.4161/rna.24604. Epub 2013 Apr 15. RNA Biol. 2013. PMID: 23696037 Free PMC article. Review.
-
Regulation of mammary epithelial cell homeostasis by lncRNAs.Int J Biochem Cell Biol. 2014 Sep;54:318-30. doi: 10.1016/j.biocel.2014.03.012. Epub 2014 Mar 26. Int J Biochem Cell Biol. 2014. PMID: 24680897 Free PMC article. Review.
-
c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer.Tumour Biol. 2016 Mar;37(3):4007-15. doi: 10.1007/s13277-015-4185-5. Epub 2015 Oct 20. Tumour Biol. 2016. PMID: 26482621
-
Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells.RNA Biol. 2014;11(6):777-87. doi: 10.4161/rna.28828. Epub 2014 Apr 24. RNA Biol. 2014. PMID: 24824789 Free PMC article.
References
-
- Mattick J. S. BioEssays. 2003;25:930–939. - PubMed
-
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Nature. 2002;420:563–573. - PubMed
-
- Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Nature. 2002;420:520–562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

