The stacked-X DNA Holliday junction and protein recognition
- PMID: 16575941
- PMCID: PMC4537160
- DOI: 10.1002/jmr.765
The stacked-X DNA Holliday junction and protein recognition
Abstract
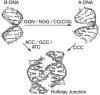
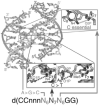
The crystal structure of the four-stranded DNA Holliday junction has now been determined in the presence and absence of junction binding proteins, with the extended open-X form of the junction seen in all protein complexes, but the more compact stacked-X structure observed in free DNA. The structures of the stacked-X junction were crystallized because of an unexpected sequence dependence on the stability of this structure. Inverted repeat sequences that contain the general motif NCC or ANC favor formation of stacked-X junctions, with the junction cross-over occurring between the first two positions of the trinucleotides. This review focuses on the sequence dependent structure of the stacked-X junction and how it may play a role in structural recognition by a class of dimeric junction resolving enzymes that themselves show no direct sequence recognition.
Copyright 2006 John Wiley & Sons, Ltd.
Figures
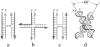


Similar articles
-
Solution formation of Holliday junctions in inverted-repeat DNA sequences.Biochemistry. 2006 Feb 28;45(8):2467-71. doi: 10.1021/bi052129x. Biochemistry. 2006. PMID: 16489738 Free PMC article.
-
The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions.Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):3971-6. doi: 10.1073/pnas.97.8.3971. Proc Natl Acad Sci U S A. 2000. PMID: 10760268 Free PMC article.
-
The structural basis of Holliday junction resolution by T7 endonuclease I.Nature. 2007 Oct 4;449(7162):621-4. doi: 10.1038/nature06158. Epub 2007 Sep 16. Nature. 2007. PMID: 17873858
-
The interaction of four-way DNA junctions with resolving enzymes.Biochem Soc Trans. 2010 Apr;38(2):399-403. doi: 10.1042/BST0380399. Biochem Soc Trans. 2010. PMID: 20298191 Review.
-
Structure of the Holliday junction: applications beyond recombination.Biochem Soc Trans. 2017 Oct 15;45(5):1149-1158. doi: 10.1042/BST20170048. Epub 2017 Aug 24. Biochem Soc Trans. 2017. PMID: 28842529 Review.
Cited by
-
The crystal structure of D212 from sulfolobus spindle-shaped virus ragged hills reveals a new member of the PD-(D/E)XK nuclease superfamily.J Virol. 2010 Jun;84(12):5890-7. doi: 10.1128/JVI.01663-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375162 Free PMC article.
-
Molecular modeling and structural studies of 12-mer immobile four-way DNA junction in solution.Bioinformation. 2014 Jul 22;10(7):394-400. doi: 10.6026/97320630010394. eCollection 2014. Bioinformation. 2014. PMID: 25187677 Free PMC article.
-
Interaction of HMG proteins and H1 with hybrid PNA-DNA junctions.Protein Sci. 2013 Nov;22(11):1552-62. doi: 10.1002/pro.2342. Epub 2013 Sep 18. Protein Sci. 2013. PMID: 23963921 Free PMC article.
-
Construction of DNA hemicatenanes from two small circular DNA molecules.PLoS One. 2015 Mar 23;10(3):e0119368. doi: 10.1371/journal.pone.0119368. eCollection 2015. PLoS One. 2015. PMID: 25799010 Free PMC article.
-
Holliday Junction Thermodynamics and Structure: Coarse-Grained Simulations and Experiments.Sci Rep. 2016 Mar 14;6:22863. doi: 10.1038/srep22863. Sci Rep. 2016. PMID: 26971574 Free PMC article.
References
-
- Arauzo-Bravo MJ, Fujii S, Kono H, Ahmad S, Sarai A. Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein-DNA recognition. J. Am. Chem. Soc. 2005;127(46):16074–16089. - PubMed
-
- Aymami J, Pous J, Lisgarten JN, Coll M. Crystallization and preliminary X-ray analysis of the DNA decamers d(CCGGATCCGG) and d(CCGGCGCCGG) Acta Crystallogr. D. Biol. Crystallogr. 2002;58(Pt 2):310–311. - PubMed
-
- Bennett RJ, West SC. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 1995a;252:213–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

