Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models
- PMID: 16579851
- PMCID: PMC1513606
- DOI: 10.1186/1471-2105-7-182
Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models
Abstract
Background: The binding between peptide epitopes and major histocompatibility complex proteins (MHCs) is an important event in the cellular immune response. Accurate prediction of the binding between short peptides and the MHC molecules has long been a principal challenge for immunoinformatics. Recently, the modeling of MHC-peptide binding has come to emphasize quantitative predictions: instead of categorizing peptides as "binders" or "non-binders" or as "strong binders" and "weak binders", recent methods seek to make predictions about precise binding affinities.
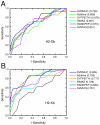
Results: We developed a quantitative support vector machine regression (SVR) approach, called SVRMHC, to model peptide-MHC binding affinities. As a non-linear method, SVRMHC was able to generate models that out-performed existing linear models, such as the "additive method". By adopting a new "11-factor encoding" scheme, SVRMHC takes into account similarities in the physicochemical properties of the amino acids constituting the input peptides. When applied to MHC-peptide binding data for three mouse class I MHC alleles, the SVRMHC models produced more accurate predictions than those produced previously. Furthermore, comparisons based on Receiver Operating Characteristic (ROC) analysis indicated that SVRMHC was able to out-perform several prominent methods in identifying strongly binding peptides.
Conclusion: As a method with demonstrated performance in the quantitative modeling of MHC-peptide binding and in identifying strong binders, SVRMHC is a promising immunoinformatics tool with not inconsiderable future potential.
Figures

Similar articles
-
In silico prediction of peptide-MHC binding affinity using SVRMHC.Methods Mol Biol. 2007;409:283-91. doi: 10.1007/978-1-60327-118-9_20. Methods Mol Biol. 2007. PMID: 18450008
-
SVRMHC prediction server for MHC-binding peptides.BMC Bioinformatics. 2006 Oct 23;7:463. doi: 10.1186/1471-2105-7-463. BMC Bioinformatics. 2006. PMID: 17059589 Free PMC article.
-
Learning MHC I--peptide binding.Bioinformatics. 2006 Jul 15;22(14):e227-35. doi: 10.1093/bioinformatics/btl255. Bioinformatics. 2006. PMID: 16873476
-
Methods and protocols for prediction of immunogenic epitopes.Brief Bioinform. 2007 Mar;8(2):96-108. doi: 10.1093/bib/bbl038. Epub 2006 Oct 31. Brief Bioinform. 2007. PMID: 17077136 Review.
-
MHC class I/peptide interactions: binding specificity and kinetics.J Mol Recognit. 1993 Jun;6(2):59-69. doi: 10.1002/jmr.300060204. J Mol Recognit. 1993. PMID: 8305252 Review.
Cited by
-
Accurate prediction of immunogenic T-cell epitopes from epitope sequences using the genetic algorithm-based ensemble learning.PLoS One. 2015 May 28;10(5):e0128194. doi: 10.1371/journal.pone.0128194. eCollection 2015. PLoS One. 2015. PMID: 26020952 Free PMC article.
-
Comparison of the predicted population coverage of tuberculosis vaccine candidates Ag85B-ESAT-6, Ag85B-TB10.4, and Mtb72f via a bioinformatics approach.PLoS One. 2012;7(7):e40882. doi: 10.1371/journal.pone.0040882. Epub 2012 Jul 17. PLoS One. 2012. PMID: 22815851 Free PMC article.
-
Peptide binding prediction for the human class II MHC allele HLA-DP2: a molecular docking approach.BMC Struct Biol. 2011 Jul 14;11:32. doi: 10.1186/1472-6807-11-32. BMC Struct Biol. 2011. PMID: 21752305 Free PMC article.
-
POPISK: T-cell reactivity prediction using support vector machines and string kernels.BMC Bioinformatics. 2011 Nov 15;12:446. doi: 10.1186/1471-2105-12-446. BMC Bioinformatics. 2011. PMID: 22085524 Free PMC article.
-
Prediction of supertype-specific HLA class I binding peptides using support vector machines.J Immunol Methods. 2007 Mar 30;320(1-2):143-54. doi: 10.1016/j.jim.2006.12.011. Epub 2007 Jan 25. J Immunol Methods. 2007. PMID: 17303158 Free PMC article.
References
-
- Flower DR, Doytchinova IA, Paine K, P. T, Blythe MJ, Lamponi D, Zygouri C, Guan P, McSparron H, H. K. Computational Vaccine Design. In: Flower DR, editor. Drug Design: Cutting Edge Approaches. Cambridge , Royal Society of Chemisty; 2002. pp. 136–180.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

