Protein stability promotes evolvability
- PMID: 16581913
- PMCID: PMC1458665
- DOI: 10.1073/pnas.0510098103
Protein stability promotes evolvability
Abstract
The biophysical properties that enable proteins to so readily evolve to perform diverse biochemical tasks are largely unknown. Here, we show that a protein's capacity to evolve is enhanced by the mutational robustness conferred by extra stability. We use simulations with model lattice proteins to demonstrate how extra stability increases evolvability by allowing a protein to accept a wider range of beneficial mutations while still folding to its native structure. We confirm this view experimentally by mutating marginally stable and thermostable variants of cytochrome P450 BM3. Mutants of the stabilized parent were more likely to exhibit new or improved functions. Only the stabilized P450 parent could tolerate the highly destabilizing mutations needed to confer novel activities such as hydroxylating the antiinflammatory drug naproxen. Our work establishes a crucial link between protein stability and evolution. We show that we can exploit this link to discover protein functions, and we suggest how natural evolution might do the same.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


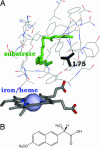
References
-
- Kauffman S. A. The Origins of Order: Self-Organization and Selection in Evolution. Oxford: Oxford Univ. Press; 1993.
-
- Bloom J. D., Meyer M. M., Meinhold P., Otey C. R., MacMillan D., Arnold F. H. Curr. Opin. Struct. Biol. 2005;15:447–452. - PubMed
-
- Kicinger R., Arciszewski T., Jong K. D. Computers Struct. 2005;83:1943–1978.
-
- Aharoni A., Gaidukov L., Khersonsky O., Gould S. M., Roodveldt C., Tawfik D. S. Nat. Genet. 2005;37:73–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

