Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis
- PMID: 16582011
- PMCID: PMC1456861
- DOI: 10.1105/tpc.105.036913
Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis
Abstract
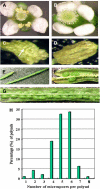
To investigate how and when sister chromatid cohesion is released from chromosomes in plants, we isolated the Arabidopsis thaliana homolog of separase (AESP) and investigated its role in somatic and meiotic cells. AESP is similar to separase proteins identified in other organisms but contains several additional structural motifs. The characterization of two Arabidopsis T-DNA insertion alleles for AESP demonstrated that it is an essential gene. Seeds homozygous for T-DNA insertions in AESP exhibited embryo arrest at the globular stage. The endosperm also exhibited a weak titan-like phenotype. Transgenic plants expressing AESP RNA interference (RNAi) from the meiosis-specific DMC1 promoter exhibited alterations in chromosome segregation during meiosis I and II that resulted in polyads containing from one to eight microspores. Consistent with its predicted role in the release of sister chromatid cohesion, immunolocalization studies showed that the removal of SYN1 from chromosome arms and the centromeres is inhibited in the RNAi mutants. However, the release of SYN1 during diplotene occurred normally, indicating that this process is independent of AESP. Therefore, our results demonstrate that AESP plays an essential role in embryo development and provide direct evidence that AESP is required for the removal of cohesin from meiotic chromosomes.
Figures



Similar articles
-
Arabidopsis separase functions beyond the removal of sister chromatid cohesion during meiosis.Plant Physiol. 2009 Sep;151(1):323-33. doi: 10.1104/pp.109.140699. Epub 2009 Jul 10. Plant Physiol. 2009. PMID: 19592426 Free PMC article.
-
The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in Arabidopsis.Plant J. 2009 Jul;59(1):1-13. doi: 10.1111/j.1365-313X.2009.03845.x. Epub 2009 Feb 18. Plant J. 2009. PMID: 19228337
-
SHUGOSHINs and PATRONUS protect meiotic centromere cohesion in Arabidopsis thaliana.Plant J. 2014 Mar;77(5):782-94. doi: 10.1111/tpj.12432. Epub 2014 Feb 12. Plant J. 2014. PMID: 24506176
-
Chromosome cohesion - rings, knots, orcs and fellowship.J Cell Sci. 2008 Jul 1;121(Pt 13):2107-14. doi: 10.1242/jcs.029132. J Cell Sci. 2008. PMID: 18565823 Review.
-
Keeping sister chromatids together: cohesins in meiosis.Reproduction. 2005 Dec;130(6):783-90. doi: 10.1530/rep.1.00864. Reproduction. 2005. PMID: 16322538 Review.
Cited by
-
The Role of Anaphase-Promoting Complex/Cyclosome (APC/C) in Plant Reproduction.Front Plant Sci. 2021 Feb 24;12:642934. doi: 10.3389/fpls.2021.642934. eCollection 2021. Front Plant Sci. 2021. PMID: 33719322 Free PMC article. Review.
-
The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis.BMC Plant Biol. 2008 Nov 28;8:121. doi: 10.1186/1471-2229-8-121. BMC Plant Biol. 2008. PMID: 19040736 Free PMC article.
-
Arabidopsis separase functions beyond the removal of sister chromatid cohesion during meiosis.Plant Physiol. 2009 Sep;151(1):323-33. doi: 10.1104/pp.109.140699. Epub 2009 Jul 10. Plant Physiol. 2009. PMID: 19592426 Free PMC article.
-
Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis.Plant Physiol. 2013 Sep;163(1):431-40. doi: 10.1104/pp.113.221713. Epub 2013 Jul 22. Plant Physiol. 2013. PMID: 23878078 Free PMC article.
-
Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes.BMC Plant Biol. 2010 Dec 17;10:280. doi: 10.1186/1471-2229-10-280. BMC Plant Biol. 2010. PMID: 21167045 Free PMC article.
References
-
- An, Y., McDowell, J., Huang, S., McKinney, E., Chambliss, S., and Meagher, R. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. - PubMed
-
- Armstrong, S., and Jones, G. (2003). Meiotic cytology and chromosome behavior in wild-type Arabidopsis thaliana. J. Exp. Bot. 54 1–10. - PubMed
-
- Barrett, A.J., and Rawlings, N.D. (2001). Evolutionary lines of cysteine peptidases. Biol. Chem. 382 727–733. - PubMed
-
- Bhatt, A.M., Lister, C., Page, T., Fransz, P., Findlay, K., Jones, G.H., Dickinson, H.G., and Dean, C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19 463–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

