Genomic analysis of the Opi- phenotype
- PMID: 16582425
- PMCID: PMC1526532
- DOI: 10.1534/genetics.106.057489
Genomic analysis of the Opi- phenotype
Abstract
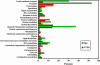
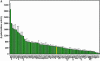
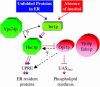
Most of the phospholipid biosynthetic genes of Saccharomyces cerevisiae are coordinately regulated in response to inositol and choline. Inositol affects the intracellular levels of phosphatidic acid (PA). Opi1p is a repressor of the phospholipid biosynthetic genes and specifically binds PA in the endoplasmic reticulum. In the presence of inositol, PA levels decrease, releasing Opi1p into the nucleus where it represses transcription. The opi1 mutant overproduces and excretes inositol into the growth medium in the absence of inositol and choline (Opi(-) phenotype). To better understand the mechanism of Opi1p repression, the viable yeast deletion set was screened to identify Opi(-) mutants. In total, 89 Opi(-) mutants were identified, of which 7 were previously known to have the Opi(-) phenotype. The Opi(-) mutant collection included genes with roles in phospholipid biosynthesis, transcription, protein processing/synthesis, and protein trafficking. Included in this set were all nonessential components of the NuA4 HAT complex and six proteins in the Rpd3p-Sin3p HDAC complex. It has previously been shown that defects in phosphatidylcholine synthesis (cho2 and opi3) yield the Opi(-) phenotype because of a buildup of PA. However, in this case the Opi(-) phenotype is conditional because PA can be shuttled through a salvage pathway (Kennedy pathway) by adding choline to the growth medium. Seven new mutants present in the Opi(-) collection (fun26, kex1, nup84, tps1, mrpl38, mrpl49, and opi10/yol032w) were also suppressed by choline, suggesting that these affect PC synthesis. Regulation in response to inositol is also coordinated with the unfolded protein response (UPR). Consistent with this, several Opi(-) mutants were found to affect the UPR (yhi9, ede1, and vps74).
Figures





Similar articles
-
Identification of novel dominant INO2c mutants with an Opi- phenotype.Mol Microbiol. 2004 Jun;52(5):1271-80. doi: 10.1111/j.1365-2958.2004.04069.x. Mol Microbiol. 2004. PMID: 15165231
-
Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor.Mol Microbiol. 2005 May;56(3):696-707. doi: 10.1111/j.1365-2958.2004.04499.x. Mol Microbiol. 2005. PMID: 15819625
-
Transcription regulation of the Saccharomyces cerevisiae PIS1 gene by inositol and the pleiotropic regulator, Ume6p.Mol Microbiol. 2008 Dec;70(6):1529-39. doi: 10.1111/j.1365-2958.2008.06506.x. Epub 2008 Oct 23. Mol Microbiol. 2008. PMID: 19019152
-
Phospholipid synthesis in yeast: regulation by phosphorylation.Biochem Cell Biol. 2004 Feb;82(1):62-70. doi: 10.1139/o03-064. Biochem Cell Biol. 2004. PMID: 15052328 Review.
-
Orchestrating phospholipid biosynthesis: Phosphatidic acid conducts and Opi1p performs.J Biol Chem. 2017 Nov 10;292(45):18729-18730. doi: 10.1074/jbc.H117.809970. J Biol Chem. 2017. PMID: 29127205 Free PMC article. Review.
Cited by
-
Systematic genetic array analysis links the Saccharomyces cerevisiae SAGA/SLIK and NuA4 component Tra1 to multiple cellular processes.BMC Genet. 2008 Jul 10;9:46. doi: 10.1186/1471-2156-9-46. BMC Genet. 2008. PMID: 18616809 Free PMC article.
-
Vitamin requirements and biosynthesis in Saccharomyces cerevisiae.Yeast. 2020 Apr;37(4):283-304. doi: 10.1002/yea.3461. Epub 2020 Feb 6. Yeast. 2020. PMID: 31972058 Free PMC article. Review.
-
Cfs1p, a Novel Membrane Protein in the PQ-Loop Family, Is Involved in Phospholipid Flippase Functions in Yeast.G3 (Bethesda). 2017 Jan 5;7(1):179-192. doi: 10.1534/g3.116.035238. G3 (Bethesda). 2017. PMID: 28057802 Free PMC article.
-
Filament formation in Saccharomyces cerevisiae--a review.Folia Microbiol (Praha). 2008;53(1):3-14. doi: 10.1007/s12223-008-0001-6. Epub 2008 May 15. Folia Microbiol (Praha). 2008. PMID: 18481212 Review.
-
N88S seipin-related seipinopathy is a lipidopathy associated with loss of iron homeostasis.Cell Commun Signal. 2025 Jan 7;23(1):10. doi: 10.1186/s12964-024-02007-9. Cell Commun Signal. 2025. PMID: 39773523 Free PMC article.
References
-
- Ambroziak, J., and S. A. Henry, 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269: 15344–15349. - PubMed
-
- Bowers, K., and T. H. Stevens, 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744: 438–454. - PubMed
-
- Carman, G. M., and S. A. Henry, 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38: 361–399. - PubMed
-
- Carrozza, M. J., R. T. Utley, J. L. Workman and J. Coté, 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19: 321–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

