N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression
- PMID: 16582428
- PMCID: PMC1526498
- DOI: 10.1534/genetics.106.056820
N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression
Abstract
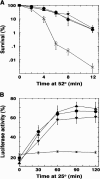
Hsp104 is a hexameric protein chaperone that resolubilizes stress-damaged proteins from aggregates. Hsp104 promotes [PSI(+)] prion propagation by breaking prion aggregates, which propagate as amyloid fibers, into more numerous prion "seeds." Inactivating Hsp104 cures cells of [PSI(+)] and other amyloid-like yeast prions. Overexpressing Hsp104 also eliminates [PSI(+)], presumably by completely resolubilizing prion aggregates. Inexplicably, however, excess Hsp104 does not cure the other prions. Here we identify missense mutations in Hsp104's amino-terminal domain (NTD), which is conserved among Hsp100 proteins but whose function is unknown, that improve [PSI(+)] propagation. Hsp104Delta147, engineered to lack the NTD, supported [PSI(+)] and functioned normally in thermotolerance and protein disaggregation. Hsp104Delta147 failed to cure [PSI(+)] when overexpressed, however, implying that excess Hsp104 does not eliminate [PSI(+)] by direct dissolution of prion aggregates. Curing of [PSI(+)] by overexpressing catalytically inactive Hsp104 (Hsp104KT), which interferes with endogenous Hsp104, did not require the NTD. We further found that Hsp104 mutants defective in threading peptides through the hexamer pore had reduced ability to support [PSI(+)] in proportion to protein resolubilization defects, suggesting that [PSI(+)] propagation depends on this threading and that Hsp104 "breaks" prion aggregates by extracting protein monomers from the amyloid fibers.
Figures





References
-
- Barnett, M. E., M. Nagy, S. Kedzierska and M. Zolkiewski, 2005. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 280: 34940–34945. - PubMed
-
- Beinker, P., S. Schlee, Y. Groemping, R. Seidel and J. Reinstein, 2002. The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 277: 47160–47166. - PubMed
-
- Cashikar, A. G., E. C. Schirmer, D. A. Hattendorf, J. R. Glover, M. S. Ramakrishnan et al., 2002. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9: 751–760. - PubMed
-
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor. [psi+] Science 268: 880–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

