Ranked prediction of p53 targets using hidden variable dynamic modeling
- PMID: 16584535
- PMCID: PMC1557743
- DOI: 10.1186/gb-2006-7-3-r25
Ranked prediction of p53 targets using hidden variable dynamic modeling
Abstract
Full exploitation of microarray data requires hidden information that cannot be extracted using current analysis methodologies. We present a new approach, hidden variable dynamic modeling (HVDM), which derives the hidden profile of a transcription factor from time series microarray data, and generates a ranked list of predicted targets. We applied HVDM to the p53 network, validating predictions experimentally using small interfering RNA. HVDM can be applied in many systems biology contexts to predict regulation of gene activity quantitatively.
Figures
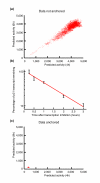
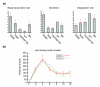
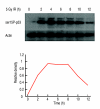
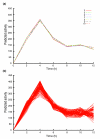
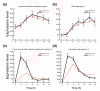
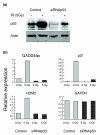
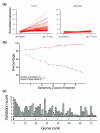
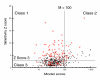
References
-
- Stark J, Brewer D, Barenco M, Tomescu D, Callard R, Hubank M. Reconstructing gene networks: what are the limits? Biochem Soc Trans. 2003;31:1519–1525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

