The action of selection on codon bias in the human genome is related to frequency, complexity, and chronology of amino acids
- PMID: 16584540
- PMCID: PMC1456966
- DOI: 10.1186/1471-2164-7-67
The action of selection on codon bias in the human genome is related to frequency, complexity, and chronology of amino acids
Abstract
Background: The question of whether synonymous codon choice is affected by cellular tRNA abundance has been positively answered in many organisms. In some recent works, concerning the human genome, this relation has been studied, but no conclusive answers have been found. In the human genome, the variation in base composition and the absence of cellular tRNA count data makes the study of the question more complicated. In this work we study the relation between codon choice and tRNA abundance in the human genome by correcting relative codon usage for background base composition and using a measure based on tRNA-gene copy numbers as a rough estimate of tRNA abundance.
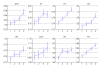
Results: We term major codons to be those codons with a relatively large tRNA-gene copy number for their corresponding amino acid. We use two measures of expression: breadth of expression (the number of tissues in which a gene was expressed) and maximum expression level among tissues (the highest value of expression of a gene among tissues). We show that for half the amino acids in the study (8 of 16) the relative major codon usage rises with breadth of expression. We show that these amino acids are significantly more frequent, are smaller and simpler, and are more ancient than the rest of the amino acids. Similar, although weaker, results were obtained for maximum expression level.
Conclusion: There is evidence that codon bias in the human genome is related to selection, although the selection forces acting on codon bias may not be straightforward and may be different for different amino acids. We suggest that, in the first group of amino acids, selection acts to enhance translation efficiency in highly expressed genes by preferring major codons, and acts to reduce translation rate in lowly expressed genes by preferring non-major ones. In the second group of amino acids other selection forces, such as reducing misincorporation rate of expensive amino acids, in terms of their size/complexity, may be in action. The fact that codon usage is more strongly related to breadth of expression than to maximum expression level supports the notion, presented in a recent study, that codon choice may be related to the tRNA abundance in the tissue in which a gene is expressed.
Figures

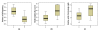
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

