Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate
- PMID: 16584714
- PMCID: PMC10561171
- DOI: 10.1016/j.carres.2006.03.004
Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate
Abstract
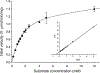
A highly sensitive fluorogenic hexosaminidase substrate, fluorescein di(N-acetyl-beta-D-glucosaminide) (FDGlcNAc), was prepared essentially as described previously [Chem. Pharm. Bull. 1993, 41, 314] with some modifications. The fluorescent analog is a substrate for a number of hexosaminidases but here we have focused on the cytoplasmic O-GlcNAcase isoforms. Kinetic analysis using purified O-GlcNAcase and its splice variant (v-O-GlcNAcase) expressed in Escherichia coli suggests that FDGlcNAc is a much more efficient substrate (Km = 84.9 microM) than the conventional substrate, para-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside (pNP-beta-GlcNAc, Km = 1.1 mM) and a previously developed fluorogenic substrate, 4-methylumbelliferyl 2-acetamido-2-deoxy-beta-D-glucopyranoside [MUGlcNAc, Km = 0.43 mM; J. Biol. Chem. 2005, 280, 25313] for O-GlcNAcase. The variant O-GlcNAcase, a protein lacking the C-terminal third of the full-length O-GlcNAcase, exhibited a Km of 2.1 mM with respect to FDGlcNAc. This shorter isoform was not previously thought to exhibit O-GlcNAcase activity based on in vitro studies with pNP-beta-GlcNAc. However, both O-GlcNAcase isoforms reduced O-GlcNAc protein levels extracted from HeLa and HT-29 cells in vitro, indicating that the splice variant is a bona fide O-GlcNAcase. Fluorescein di-N-acetyl-beta-D-galactosaminide (FDGalNAc) is not cleaved by these enzymes, consistent with previous findings that the O-GlcNAcase has substrate specificity toward O-GlcNAc but not O-GalNAc. The enzymatic activity of the shorter isoform of O-GlcNAcase was first detected by using highly sensitive fluorogenic FDGlcNAc substrate. The finding that O-GlcNAcase exists as two distinct isoforms has a number of important implications for the role of O-GlcNAcase in hexosamine signaling.
Figures




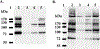


Similar articles
-
Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity.Nat Struct Mol Biol. 2006 Apr;13(4):365-71. doi: 10.1038/nsmb1079. Epub 2006 Mar 26. Nat Struct Mol Biol. 2006. PMID: 16565725
-
Location and characterization of the O-GlcNAcase active site.Biochim Biophys Acta. 2006 May;1760(5):829-39. doi: 10.1016/j.bbagen.2006.01.017. Epub 2006 Feb 20. Biochim Biophys Acta. 2006. PMID: 16517082
-
Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants.Biochemistry. 2006 Mar 21;45(11):3835-44. doi: 10.1021/bi052370b. Biochemistry. 2006. PMID: 16533067
-
The hexosamine signaling pathway: deciphering the "O-GlcNAc code".Sci STKE. 2005 Nov 29;2005(312):re13. doi: 10.1126/stke.3122005re13. Sci STKE. 2005. PMID: 16317114 Review.
-
In Vitro Biochemical Assays for O-GlcNAc-Processing Enzymes.Chembiochem. 2017 Aug 4;18(15):1462-1472. doi: 10.1002/cbic.201700138. Epub 2017 Jun 19. Chembiochem. 2017. PMID: 28474822 Review.
Cited by
-
Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis.Cell Stress Chaperones. 2013 Sep;18(5):535-58. doi: 10.1007/s12192-013-0426-y. Epub 2013 Apr 26. Cell Stress Chaperones. 2013. PMID: 23620203 Free PMC article.
-
Short O-GlcNAcase Is Targeted to the Mitochondria and Regulates Mitochondrial Reactive Oxygen Species Level.Cells. 2022 Jun 2;11(11):1827. doi: 10.3390/cells11111827. Cells. 2022. PMID: 35681522 Free PMC article.
-
Increased O-GlcNAc levels correlate with decreased O-GlcNAcase levels in Alzheimer disease brain.Biochim Biophys Acta. 2014 Sep;1842(9):1333-9. doi: 10.1016/j.bbadis.2014.05.014. Epub 2014 May 23. Biochim Biophys Acta. 2014. PMID: 24859566 Free PMC article.
-
Recognition of diazirine-modified O-GlcNAc by human O-GlcNAcase.Medchemcomm. 2014 Aug 1;5(8):1227-1234. doi: 10.1039/C4MD00164H. Medchemcomm. 2014. PMID: 25068034 Free PMC article.
-
Chemical tools to explore nutrient-driven O-GlcNAc cycling.Crit Rev Biochem Mol Biol. 2014 Jul-Aug;49(4):327-42. doi: 10.3109/10409238.2014.931338. Crit Rev Biochem Mol Biol. 2014. PMID: 25039763 Free PMC article. Review.
References
-
- Jackson SP; Tjian R. Cell 1988, 55, 125–133. - PubMed
-
- Kelly WG; Dahmus ME; Hart GW J. Biol. Chem 1993, 268, 10416–10424. - PubMed
-
- Holt GW; Haltiwanger RS; Torres C-R; Hart GW J. Biol. Chem 1987, 262, 14847–14850. - PubMed
-
- Dong L-Y; Xu Z-S; Chevrier MR; Cotter RJ;Cleveland DW; Hart GW J. Biol. Chem 1993, 268, 16679–16687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

