Crystal structure of the Anopheles gambiae 3-hydroxykynurenine transaminase
- PMID: 16585514
- PMCID: PMC1458638
- DOI: 10.1073/pnas.0510233103
Crystal structure of the Anopheles gambiae 3-hydroxykynurenine transaminase
Abstract
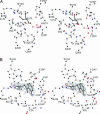
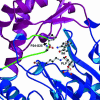
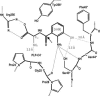
In Anopheles gambiae, the vector for the most deadly malaria parasite Plasmodium falciparum, xanthurenic acid (XA) plays a key role in parasite gametogenesis and fertility. In mosquitoes, XA is produced by transamination of 3-hydroxykynurenine (3-HK), a reaction that represents the main route to prevent the accumulation of the potentially toxic 3-HK excess. Interfering with XA metabolism in A. gambiae therefore appears an attractive avenue for the development of malaria transmission-blocking drugs and insecticides. We have determined the crystal structure of A. gambiae 3-HK transaminase in its pyridoxal 5'-phosphate form and in complex with a newly synthesized competitive enzyme inhibitor. Structural inspection of the enzyme active site reveals the key molecular determinants for ligand recognition and catalysis. Major contributions toward inhibitor binding are provided by a salt bridge between the inhibitor carboxylate and Arg-356 and by a remarkable hydrogen bond network involving the anthranilic moiety of the inhibitor and backbone atoms of residues Gly-25 and Asn-44. This study may be useful for the structure-based design of specific enzyme inhibitors of potential interest as antimalarial agents.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
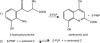
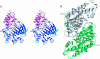

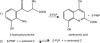
Similar articles
-
Identification and biochemical characterization of the Anopheles gambiae 3-hydroxykynurenine transaminase.FEBS J. 2005 Nov;272(21):5653-62. doi: 10.1111/j.1742-4658.2005.04961.x. FEBS J. 2005. PMID: 16262702
-
Study of Anopheles gambiae 3-hydroxykynurenine transaminase activity and inhibition by LC-MS/MS method.J Pharm Biomed Anal. 2019 Sep 5;173:154-161. doi: 10.1016/j.jpba.2019.05.025. Epub 2019 May 13. J Pharm Biomed Anal. 2019. PMID: 31129535
-
Crystal structures of Aedes aegypti alanine glyoxylate aminotransferase.J Biol Chem. 2006 Dec 1;281(48):37175-82. doi: 10.1074/jbc.M607032200. Epub 2006 Sep 21. J Biol Chem. 2006. PMID: 16990263
-
[Malaria: from genetic and molecular biology to disease control].J Soc Biol. 2004;198(3):181-5. J Soc Biol. 2004. PMID: 15662933 Review. French.
-
Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complement-like pathway.Cell Host Microbe. 2008 Jun 12;3(6):364-74. doi: 10.1016/j.chom.2008.05.007. Cell Host Microbe. 2008. PMID: 18541213 Review.
Cited by
-
Evolution of two alanine glyoxylate aminotransferases in mosquito.Biochem J. 2006 Aug 1;397(3):473-81. doi: 10.1042/BJ20060469. Biochem J. 2006. PMID: 16681462 Free PMC article.
-
Biochemical Evolution of a Potent Target of Mosquito Larvicide, 3-Hydroxykynurenine Transaminase.Molecules. 2022 Aug 2;27(15):4929. doi: 10.3390/molecules27154929. Molecules. 2022. PMID: 35956879 Free PMC article.
-
A second generation of 1,2,4-oxadiazole derivatives with enhanced solubility for inhibition of 3-hydroxykynurenine transaminase (HKT) from Aedes aegypti.RSC Med Chem. 2020 Dec 9;12(2):222-236. doi: 10.1039/d0md00305k. eCollection 2021 Mar 4. RSC Med Chem. 2020. PMID: 34046611 Free PMC article.
-
Insecticidal activity of indole derivatives against Plutella xylostella and selectivity to four non-target organisms.Ecotoxicology. 2019 Oct;28(8):973-982. doi: 10.1007/s10646-019-02095-1. Epub 2019 Aug 16. Ecotoxicology. 2019. PMID: 31420785
-
On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination.Acta Crystallogr D Biol Crystallogr. 2009 Oct;65(Pt 10):1089-97. doi: 10.1107/S0907444909029643. Epub 2009 Sep 16. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19770506 Free PMC article.
References
-
- Stone T. W. Toxicon. 2001;39:61–73. - PubMed
-
- Schwarcz R., Pellicciari R. J. Pharmacol. Exp. Ther. 2002;1:1–10. - PubMed
-
- Breton J., Avanzi N., Magagnin S., Covini N., Magistrelli G., Cozzi L., Isacchi A. Eur. J. Biochem. 2000;267:1092–1099. - PubMed
-
- Hirai M., Kiuchi M., Wang J., Ishii A., Matsuoka H. Insect Mol. Biol. 2002;11:497–504. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

