Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection
- PMID: 16585524
- PMCID: PMC1458687
- DOI: 10.1073/pnas.0601523103
Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection
Abstract
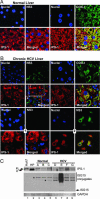
Viral signaling through retinoic acid-inducible gene-I (RIG-I) and its adaptor protein, IFN promoter-stimulator 1 (IPS-1), activates IFN regulatory factor-3 (IRF-3) and the host IFN-alpha/beta response that limits virus infection. The hepatitis C virus (HCV) NS3/4A protease cleaves IPS-1 to block RIG-I signaling, but how this regulation controls the host response to HCV is not known. Moreover, endogenous IPS-1 cleavage has not been demonstrated in the context of HCV infection in vitro or in vivo. Here, we show that HCV infection transiently induces RIG-I- and IPS-1-dependent IRF-3 activation. This host response limits HCV production and constrains cellular permissiveness to infection. However, HCV disrupts this response early in infection by NS3/4A cleavage of IPS-1 at C508, releasing IPS-1 from the mitochondrial membrane. Cleavage results in subcellular redistribution of IPS-1 and loss of interaction with RIG-I, thereby preventing downstream activation of IRF-3 and IFN-beta induction. Liver tissues from chronically infected patients similarly demonstrate subcellular redistribution of IPS-1 in infected hepatocytes and IPS-1 cleavage associated with a lack of ISG15 expression and conjugation of target proteins in vivo. Importantly, small-molecule inhibitors of NS3/4A prevent cleavage and restore RIG-I signaling of IFN-beta induction. Our results suggest a dynamic model in which early activation of IRF-3 and induction of antiviral genes are reversed by IPS-1 proteolysis and abrogation of RIG-I signaling as NS3/4A accumulates in newly infected cells. HCV protease inhibitors effectively prevent IPS-1 proteolysis, suggesting they may be capable of restoring this innate host response in clinical practice.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


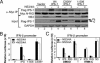
Similar articles
-
Restoration of the activated Rig-I pathway in hepatitis C virus (HCV) replicon cells by HCV protease, polymerase, and NS5A inhibitors in vitro at clinically relevant concentrations.Antimicrob Agents Chemother. 2013 Sep;57(9):4417-26. doi: 10.1128/AAC.00399-13. Epub 2013 Jul 8. Antimicrob Agents Chemother. 2013. PMID: 23836176 Free PMC article.
-
Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense.J Biol Chem. 2007 Apr 6;282(14):10792-803. doi: 10.1074/jbc.M610361200. Epub 2007 Feb 8. J Biol Chem. 2007. PMID: 17289677
-
Hepatitis C virus NS2 and NS3/4A proteins are potent inhibitors of host cell cytokine/chemokine gene expression.Virol J. 2006 Sep 1;3:66. doi: 10.1186/1743-422X-3-66. Virol J. 2006. PMID: 16945160 Free PMC article.
-
[Innate immune responses against viral infection and its suppression by viral proteins].Yakugaku Zasshi. 2013;133(3):323-8. doi: 10.1248/yakushi.12-00237-5. Yakugaku Zasshi. 2013. PMID: 23449408 Review. Japanese.
-
Innate immunity and HCV.J Hepatol. 2013 Mar;58(3):564-74. doi: 10.1016/j.jhep.2012.10.005. Epub 2012 Oct 11. J Hepatol. 2013. PMID: 23063572 Review.
Cited by
-
Isoflavone agonists of IRF-3 dependent signaling have antiviral activity against RNA viruses.J Virol. 2012 Jul;86(13):7334-44. doi: 10.1128/JVI.06867-11. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532686 Free PMC article.
-
Two new monoclonal antibodies for biochemical and flow cytometric analyses of human interferon regulatory factor-3 activation, turnover, and depletion.Methods. 2013 Feb;59(2):225-32. doi: 10.1016/j.ymeth.2012.05.011. Epub 2012 Jun 13. Methods. 2013. PMID: 22705311 Free PMC article.
-
Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction.J Virol. 2010 Feb;84(4):1785-91. doi: 10.1128/JVI.01362-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007272 Free PMC article.
-
Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins.Proc Natl Acad Sci U S A. 2007 Apr 3;104(14):5848-53. doi: 10.1073/pnas.0700760104. Epub 2007 Mar 21. Proc Natl Acad Sci U S A. 2007. PMID: 17376867 Free PMC article.
-
Toll-like Receptor Response to Hepatitis C Virus Infection: A Recent Overview.Int J Mol Sci. 2022 May 13;23(10):5475. doi: 10.3390/ijms23105475. Int J Mol Sci. 2022. PMID: 35628287 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI060389/AI/NIAID NIH HHS/United States
- R01AI060389/AI/NIAID NIH HHS/United States
- U19 AI040035/AI/NIAID NIH HHS/United States
- U01 AI048235/AI/NIAID NIH HHS/United States
- R01 AA012863/AA/NIAAA NIH HHS/United States
- R21DA018054/DA/NIDA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 DK068598/DK/NIDDK NIH HHS/United States
- R21 DA018054/DA/NIDA NIH HHS/United States
- R01AA012863/AA/NIAAA NIH HHS/United States
- U01AI48235/AI/NIAID NIH HHS/United States
- R01DK068598/DK/NIDDK NIH HHS/United States
- U19AI40035/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

