Morphology of isolated Gli349, a leg protein responsible for Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy
- PMID: 16585743
- PMCID: PMC1447022
- DOI: 10.1128/JB.188.8.2821-2828.2006
Morphology of isolated Gli349, a leg protein responsible for Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy
Abstract
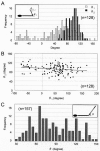
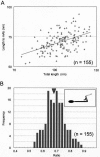
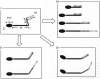
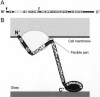
Several species of mycoplasmas rely on an unknown mechanism to glide across solid surfaces in the direction of a membrane protrusion at the cell pole. Our recent studies on the fastest species, Mycoplasma mobile, suggested that a 349-kDa protein, Gli349, localized at the base of the membrane protrusion called the neck, forms legs that stick out from the neck and propel the cell by repeatedly binding to and releasing from a solid surface, based on the energy of ATP hydrolysis. Here, the Gli349 protein was isolated from mycoplasma cells and its structure was analyzed. Gel filtration analysis showed that the isolated Gli349 protein is monomeric. Rotary shadowing electron microscopy revealed that the molecular structure resembles the symbol for an eighth note in music. It contains an oval foot 14 nm long in axis. From this foot extend three rods in tandem of 43, 20, and 20 nm, in that order. The hinge connecting the first and second rods is flexible, while the next hinge has a distinct preference in its angle, near 90 degrees. Molecular images revealed that a monoclonal antibody that can bind to the position at one-third of the total peptide length from the N terminus bound to a position two-thirds from the foot end, suggesting that the foot corresponds to the C-terminal region. The amino acid sequence was assigned to the molecular image, and the topology of the molecule in the gliding machinery is discussed.
Figures





References
-
- Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58.
-
- Arata, T. 1998. Electron microscopic observation of monomeric actin attached to a myosin head. J. Struct. Biol. 123:8-16. - PubMed
-
- Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York, N.Y.
-
- Fernandez-Patron, C., L. Castellanos-Serra, and P. Rodriguez. 1992. Reverse staining of sodium dodecyl sulfate polyacrylamide gels by imidazole-zinc salts: sensitive detection of unmodified proteins. BioTechniques 12:564-573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

