HIV protease cleaves poly(A)-binding protein
- PMID: 16594896
- PMCID: PMC1462710
- DOI: 10.1042/BJ20060108
HIV protease cleaves poly(A)-binding protein
Abstract
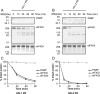
The PABP [poly(A)-binding protein] is able to interact with the 3' poly(A) tail of eukaryotic mRNA, promoting its translation. Cleavage of PABP by viral proteases encoded by several picornaviruses and caliciviruses plays a role in the abrogation of cellular protein synthesis. We report that infection of MT-2 cells with HIV-1 leads to efficient proteolysis of PABP. Analysis of PABP integrity was carried out in BHK-21 (baby-hamster kidney) and COS-7 cells upon individual expression of the protease from several members of the Retroviridae family, e.g. MoMLV (Moloney murine leukaemia virus), MMTV (mouse mammary tumour virus), HTLV-I (human T-cell leukaemia virus type I), SIV (simian immunodeficiency virus), HIV-1 and HIV-2. Moreover, protease activity against PABP was tested in a HeLa-cell-free system. Only MMTV, HIV-1 and HIV-2 proteases were able to cleave PABP in the absence of other viral proteins. Purified HIV-1 and HIV-2 proteases cleave PABP1 directly at positions 237 and 477, separating the two first RNA-recognition motifs from the C-terminal domain of PABP. An additional cleavage site located at position 410 was detected for HIV-2 protease. These findings indicate that some retroviruses may share with picornaviruses and caliciviruses the capacity to proteolyse PABP.
Figures
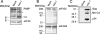




Comment in
-
Cleavage, a real turn-off? HIV-mediated proteolysis of PABP1.Biochem J. 2006 Jun 1;396(2):e9-11. doi: 10.1042/bj20060545. Biochem J. 2006. PMID: 16703665 Free PMC article.
References
-
- Hentze M. W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. - PubMed
-
- Jacobson A. Poly(A) metabolism and translation: the closed loop model. In: Hershey J. W. B., Mathews M. B., Sonenberg N., editors. Translational Control. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480.

