Potential antivirals and antiviral strategies against SARS coronavirus infections
- PMID: 16597209
- PMCID: PMC7105749
- DOI: 10.1586/14787210.4.2.291
Potential antivirals and antiviral strategies against SARS coronavirus infections
Abstract
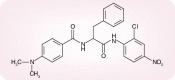
There are a number of antivirals as well as antiviral strategies that could be envisaged to prevent or treat severe acute respiratory syndrome (SARS) (or similar) coronavirus (CoV) infections. Targets for the prophylactic or therapeutic interventions include interaction of the spike (S) glycoprotein (S1 domain) with the host cell receptor, fusion of the S2 domain with the host cell membrane, processing of the replicase polyproteins by the virus-encoded proteases (3C-like cysteine protease [3CLpro] and papain-like cysteine protease) and other virus-encoded enzymes such as the NTPase/helicase and RNA-dependent RNA polymerase. Human monoclonal antibody blocking S1 may play an important role in the immunoprophylaxis of SARS. Fusion inhibitors reminiscent of enfuvirtide in the case of HIV may also be developed for SARS-CoV. Various peptidomimetic and nonpeptidic inhibitors of 3CLpro have been described, the best ones inhibiting SARS-CoV replication with a selectivity index greater than 1000. Human interferons, in particular alpha- and beta-interferon, as well as short interfering RNAs could further be pursued for the control of SARS. Various other compounds, often with an ill-defined mode of action but selectivity indexes up to 100, have been reported to exhibit in vitro activity against SARS-CoV: valinomycin, glycopeptide antibiotics, plant lectins, hesperetin, glycyrrhizin, aurintricarboxylic acid, chloroquine, niclosamide, nelfinavir and calpain inhibitors.
Figures













References
-
- Lee N, Hui D, Wu A et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994 (2003). - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003). - PubMed
-
- Drosten C, Gunther S, Preiser W et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
