16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory
- PMID: 16597863
- PMCID: PMC1448638
- DOI: 10.1128/JCM.44.4.1359-1366.2006
16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory
Abstract
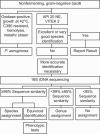
Over a period of 26 months, we have evaluated in a prospective fashion the use of 16S rRNA gene sequencing as a means of identifying clinically relevant isolates of nonfermenting gram-negative bacilli (non-Pseudomonas aeruginosa) in the microbiology laboratory. The study was designed to compare phenotypic with molecular identification. Results of molecular analyses were compared with two commercially available identification systems (API 20 NE, VITEK 2 fluorescent card; bioMérieux, Marcy l'Etoile, France). By 16S rRNA gene sequence analyses, 92% of the isolates were assigned to species level and 8% to genus level. Using API 20 NE, 54% of the isolates were assigned to species and 7% to genus level, and 39% of the isolates could not be discriminated at any taxonomic level. The respective numbers for VITEK 2 were 53%, 1%, and 46%, respectively. Fifteen percent and 43% of the isolates corresponded to species not included in the API 20 NE and VITEK 2 databases, respectively. We conclude that 16S rRNA gene sequencing is an effective means for the identification of clinically relevant nonfermenting gram-negative bacilli. Based on our experience, we propose an algorithm for proper identification of nonfermenting gram-negative bacilli in the diagnostic laboratory.
Figures
References
-
- Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Böttger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range PCR: a 3-year experience. Clin. Infect. Dis. 37:167-172. - PubMed
-
- Böttger, E. C. 1996. Approaches for identification of microorganisms. ASM News 62:247-250.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources