Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus
- PMID: 16597869
- PMCID: PMC1448641
- DOI: 10.1128/JCM.44.4.1405-1412.2006
Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus
Abstract
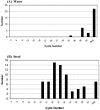
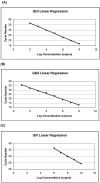
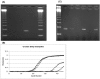
Noroviruses (NoVs) are the most commonly identified cause of outbreaks and sporadic cases of acute gastroenteritis. We evaluated and optimized NoV-specific TaqMan real-time reverse transcription (RT)-PCR assays for the rapid detection and typing of NoV strains belonging to genogroups GI and GII and adapted them to the LightCycler platform. We expanded the detection ability of the assays by developing an assay that detects the GIV NoV strain. The assays were validated with 92 clinical samples and 33 water samples from confirmed NoV outbreaks and suspected NoV contamination cases. The assays detected NoV RNA in all of the clinical specimens previously confirmed positive by conventional RT-PCR and sequencing. Additionally, the TaqMan assays successfully detected NoV RNA in water samples containing low viral concentrations and inhibitors of RT and/or PCR, whereas the conventional method with region B primers required dilution of the inhibitors. By means of serially diluted NoV T7 RNA transcripts, a potential detection limit of <10 transcript copies per reaction mixture was observed with the GII assay and a potential detection limit of <100 transcript copies per reaction mixture was observed with the GI assay. These results and the ability to detect virus in water that was negative by RT-PCR demonstrate the higher sensitivity of the TaqMan assay compared with that of a conventional RT-PCR assay. The TaqMan methods dramatically decrease the turnaround time by eliminating post-PCR processing. These assays have proven useful in assisting scientists in public health and diagnostic laboratories report findings quickly to outbreak management teams.
Figures

References
-
- Abbaszadegan, M., Lechevallier, and C. P. Gerba. 2003. Occurrence of viruses in US ground water. J. Am. Water Works Assoc. 95:107-120.
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of Norwalk-like viruses. J. Infect. Dis. 181:S336-S348. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

