Distribution of environmental mycobacteria in Karonga District, northern Malawi
- PMID: 16597928
- PMCID: PMC1449038
- DOI: 10.1128/AEM.72.4.2343-2350.2006
Distribution of environmental mycobacteria in Karonga District, northern Malawi
Abstract
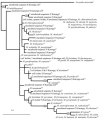
The genus Mycobacterium includes many species that are commonly found in the environment (in soil and water or associated with plants and animals), as well as species that are responsible for two major human diseases, tuberculosis (Mycobacterium tuberculosis) and leprosy (Mycobacterium leprae). The distribution of environmental mycobacteria was investigated in the context of a long-term study of leprosy, tuberculosis, Mycobacterium bovis BCG vaccination, and the responses of individuals to various mycobacterial antigens in Karonga District, northern Malawi, where epidemiological studies had indicated previously that people may be exposed to different mycobacterial species in the northern and southern parts of the district. A total of 148 soil samples and 24 water samples were collected from various locations and examined to determine the presence of mycobacteria. The detection method involved semiselective culturing and acid-fast staining, following decontamination of samples to enrich mycobacteria and reduce the numbers of other microorganisms, or PCR with primers specific for the mycobacterial 16S rRNA gene, using DNA extracted directly from soil and water samples. Mycobacteria were detected in the majority of the samples, and subsequent sequence analysis of PCR products amplified directly from soil DNA indicated that most of the products were related to known environmental mycobacteria. For both methods the rates of recovery were consistently higher for dry season samples than for wet season samples. All isolates cultured from soil appeared to be strains of Mycobacterium fortuitum. This study revealed a complex pattern for the environmental mycobacterial flora but identified no clear differences between the northern and southern parts of Karonga District.
Figures
References
-
- Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. - PubMed
-
- Black, G. F., R. E. Weir, S. D. Chaguluka, D. Warndorff, A. C. Crampin, L. Mwaungulu, L. Sichali, S. Floyd, L. Bliss, E. Jarman, L. Donovan, P. Andersen, W. Britton, G. Hewinson, K. Huygen, J. Paulsen, M. Singh, R. Prestidge, P. E. Fine, and H. M. Dockrell. 2003. Gamma interferon responses induced by a panel of recombinant and purified mycobacterial antigens in healthy, non-Mycobacterium bovis BCG-vaccinated Malawian young adults. Clin. Diagn. Lab. Immunol. 10:602-611. - PMC - PubMed
-
- Brandt, L., J. Feino Cunha, A. Weinreich Olsen, B. Chilima, P. Hirsch, R. Appelberg, and P. Andersen. 2002. Failure of the Mycobacterium bovis BCG vaccine: some species of environmental mycobacteria block multiplication of BCG and induction of protective immunity to tuberculosis. Infect. Immun. 70:672-678. - PMC - PubMed
-
- Domenech, P., M. S. Jimenez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1997. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

