Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula
- PMID: 16597930
- PMCID: PMC1448991
- DOI: 10.1128/AEM.72.4.2359-2365.2006
Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula
Abstract
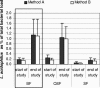
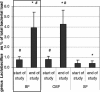
The developing intestinal microbiota of breast-fed infants is considered to play an important role in the priming of the infants' mucosal and systemic immunity. Generally, Bifidobacterium and Lactobacillus predominate the microbiota of breast-fed infants. In intervention trials it has been shown that lactobacilli can exert beneficial effects on, for example, diarrhea and atopy. However, the Lactobacillus species distribution in breast-fed or formula-fed infants has not yet been determined in great detail. For accurate enumeration of different lactobacilli, duplex 5' nuclease assays, targeted on rRNA intergenic spacer regions, were developed for Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus fermentum, Lactobacillus paracasei, Lactobacillus plantarum, Lactobacillus reuteri, and Lactobacillus rhamnosus. The designed and validated assays were used to determine the amounts of different Lactobacillus species in fecal samples of infants receiving a standard formula (SF) or a standard formula supplemented with galacto- and fructo-oligosaccharides in a 9:1 ratio (OSF). A breast-fed group (BF) was studied in parallel as a reference. During the 6-week intervention period a significant increase was shown in total percentage of fecal lactobacilli in the BF group (0.8% +/- 0.3% versus 4.1% +/- 1.5%) and the OSF group (0.8% +/- 0.3% versus 4.4% +/- 1.4%). The Lactobacillus species distribution in the OSF group was comparable to breast-fed infants, with relatively high levels of L. acidophilus, L. paracasei, and L. casei. The SF-fed infants, on the other hand, contained more L. delbrueckii and less L. paracasei compared to breast-fed infants and OSF-fed infants. An infant milk formula containing a specific mixture of prebiotics is able to induce a microbiota that closely resembles the microbiota of BF infants.
Figures
References
-
- Adams, M. R., and C. J. Hall. 1988. Growth inhibition of food-borne pathogens by lactic and acetic acids and their mixtures. Int. J. Food Sci. Technol. 23:287-292.
-
- Agostoni, C., I. Axelsson, C. Braegger, O. Goulet, B. Koletzko, K. F. Michaelsen, J. Rigo, R. Shamir, H. Szajewska, D. Turck, and L. T. Weaver. 2004. Probiotic bacteria in dietetic products for infants: a commentary by the ESPGHAN Committee on Nutrition. J. Pediatr. Gastroenterol. Nutr. 38:365-374. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bakker-Zierikzee, A., M. S. Alles, J. Knol, F. Kok, J. J. M. Tolboom, and J. G. Bindels. 2005. Effects of infant formula containing a mixture of galacto- and fructooligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 94:783-790. - PubMed
-
- Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

