Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network
- PMID: 16597941
- PMCID: PMC1449017
- DOI: 10.1128/AEM.72.4.2428-2438.2006
Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network
Abstract
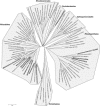
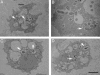
Free-living amoebae (FLA) are ubiquitous organisms that have been isolated from various domestic water systems, such as cooling towers and hospital water networks. In addition to their own pathogenicity, FLA can also act as Trojan horses and be naturally infected with amoeba-resisting bacteria (ARB) that may be involved in human infections, such as pneumonia. We investigated the biodiversity of bacteria and their amoebal hosts in a hospital water network. Using amoebal enrichment on nonnutrient agar, we isolated 15 protist strains from 200 (7.5%) samples. One thermotolerant Hartmannella vermiformis isolate harbored both Legionella pneumophila and Bradyrhizobium japonicum. By using amoebal coculture with axenic Acanthamoeba castellanii as the cellular background, we recovered at least one ARB from 45.5% of the samples. Four new ARB isolates were recovered by culture, and one of these isolates was widely present in the water network. Alphaproteobacteria (such as Rhodoplanes, Methylobacterium, Bradyrhizobium, Afipia, and Bosea) were recovered from 30.5% of the samples, mycobacteria (Mycobacterium gordonae, Mycobacterium kansasii, and Mycobacterium xenopi) were recovered from 20.5% of the samples, and Gammaproteobacteria (Legionella) were recovered from 5.5% of the samples. No Chlamydia or Chlamydia-like organisms were recovered by amoebal coculture or detected by PCR. The observed strong association between the presence of amoebae and the presence of Legionella (P < 0.001) and mycobacteria (P = 0.009) further suggests that FLA are a reservoir for these ARB and underlines the importance of considering amoebae when water control measures are designed.
Figures


References
-
- Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. - PubMed
-
- Arend, S. M., E. Cerda De Palou, P. De Haas, R. Janssen, M. A. Hoeve, E. M. Verhard, T. H. Ottenhoff, D. Van Soolingen, and J. T. Van Dissel. 2004. Pneumonia caused by Mycobacterium kansasii in a series of patients without recognised immune defect. Clin. Microbiol. Infect. 10:738-748. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

