Development and characterization of stable sediment-free anaerobic bacterial enrichment cultures that dechlorinate aroclor 1260
- PMID: 16597944
- PMCID: PMC1448987
- DOI: 10.1128/AEM.72.4.2460-2470.2006
Development and characterization of stable sediment-free anaerobic bacterial enrichment cultures that dechlorinate aroclor 1260
Abstract
We have developed sediment-free anaerobic enrichment cultures that dechlorinate a broad spectrum of highly chlorinated polychlorinated biphenyls (PCBs). The cultures were developed from Aroclor 1260-contaminated sediment from the Housatonic River in Lenox, MA. Sediment slurries were primed with 2,6-dibromobiphenyl to stimulate Process N dechlorination (primarily meta dechlorination), and sediment was gradually removed by successive transfers (10%) to minimal medium. The cultures grow on pyruvate, butyrate, or acetate plus H(2). Gas chromatography-electron capture detector analysis demonstrated that the cultures extensively dechlorinate 50 to 500 mug/ml of Aroclor 1260 at 22 to 24 degrees C by Dechlorination Process N. Triplicate cultures of the eighth transfer without sediment dechlorinated 76% of the hexa- through nonachlorobiphenyls in Aroclor 1260 (250 mug/ml) to tri- through pentachlorobiphenyls in 110 days. At least 64 PCB congeners, all of which are chlorinated on both rings and 47 of which have six or more chlorines, were substrates for this dechlorination. To characterize the bacterial diversity in the enrichments, we used eubacterial primers to amplify and clone 16S rRNA genes from DNA extracted from cultures grown on acetate plus H(2). Restriction fragment length polymorphism analysis of 107 clones demonstrated the presence of Thauera-like Betaproteobacteria, Geobacter-like Deltaproteobacteria, Pseudomonas species, various Clostridiales, Bacteroidetes, Dehalococcoides of the Chloroflexi group, and unclassified Eubacteria. Our development of highly enriched, robust, stable, sediment-free cultures that extensively dechlorinate a highly chlorinated commercial PCB mixture is a major and unprecedented breakthrough in the field. It will enable intensive study of the organisms and genes responsible for a major PCB dechlorination process that occurs in the environment and could also lead to effective remediation applications.
Figures
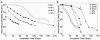



References
-
- Adrian, L., U. Szewzyk, and H. Görisch. 2000. Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation 11:73-81. - PubMed
-
- Alder, A. C., M. M. Häggblom, S. R. Oppenheimer, and L. Y. Young. 1993. Reductive dechlorination of polychlorinated biphenyls in anaerobic sediments. Environ. Sci. Technol. 27:530-538.
-
- Altschul, S. F., Y. Ma, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

