Cloning and sequencing of the ompA gene of Enterobacter sakazakii and development of an ompA-targeted PCR for rapid detection of Enterobacter sakazakii in infant formula
- PMID: 16597955
- PMCID: PMC1449048
- DOI: 10.1128/AEM.72.4.2539-2546.2006
Cloning and sequencing of the ompA gene of Enterobacter sakazakii and development of an ompA-targeted PCR for rapid detection of Enterobacter sakazakii in infant formula
Abstract
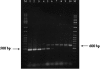
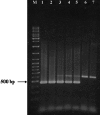
Enterobacter sakazakii is an emerging, infant formula-borne pathogen that causes severe meningitis, meningoencephalitis, sepsis, and necrotizing enterocolitis in neonates and infants, with a high fatality rate. Traditional detection methods take up to 7 days to identify E. sakazakii. The outer membrane protein A gene (ompA), along with its flanking sequences from E. sakazakii (ATCC 51329), was cloned in the pGEM-T Easy vector and sequenced. Comparison of the nucleotide and deduced amino acid sequences of the ompA gene with other sequences available in the GenBank database revealed a high degree of homology with ompA genes of other gram-negative bacteria belonging to the Enterobacteriaceae. Based on regions of the ompA gene unique to E. sakazakii, two primers were synthesized to develop and optimize an E. sakazakii-specific PCR. The PCR amplified a 469-bp DNA product from all E. sakazakii strains tested but not from other bacteria. Experiments to determine the sensitivity of the PCR indicated that it could detect as few as 10(3) CFU/ml of E. sakazakii bacteria in infant formula directly and 10(-1) CFU/ml after an 8-h enrichment step. We conclude that this PCR, combined with enrichment culturing, has the potential to be used as a rapid tool for detecting the presence of E. sakazakii in infant formula.
Figures



References
-
- Bar-Oz, B., A. Preminger, O. Peleg, C. Block, and I. Arad. 2002. Enterobacter sakazakii infection in the newborn. Acta Paediatr. 90:356-358. - PubMed
-
- FSNET. 8. November 2002, posting date. Recalled baby formula found in Colorado stores. Colorado Department of Public Health and Environment. [Online.] http://archives.foodsafetynetwork.ca/fsnet/2002/11-2002/fsnet_november_8...#RECALLED%20BABY.
-
- Guillaume-Gentil, O., V. Sonnard, M. C. Kandhai, J. D. Marugg, and H. Joosten. 2005. A simple and rapid cultural method for detection of Enterobacter sakazakii in environmental samples. J. Food Prot. 68:64-69. - PubMed
-
- Himelright, I., E. Harris, V. Lorch, M. Anderson, T. Jones, A. Craig, M. Kuehnert, T. Forster, M. Arduino, B. Jensen, and D. Jernigan. 2002. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. Morbid. Mortal. Wkly. Rep. 51:297-300. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

