Clonal dissemination of Escherichia coli O157:H7 subtypes among dairy farms in northeast Ohio
- PMID: 16597966
- PMCID: PMC1449053
- DOI: 10.1128/AEM.72.4.2621-2626.2006
Clonal dissemination of Escherichia coli O157:H7 subtypes among dairy farms in northeast Ohio
Abstract
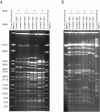
To ascertain the extent to which indistinguishable strains of Escherichia coli O157:H7 are shared between farms, molecular characterization was performed on E. coli O157:H7 isolates recovered during a longitudinal study of 20 dairy farms in northeast Ohio. Of the 20 dairy farms sampled, 16 were located in a primary area and 4 were located in two other distant geographical areas. A total of 92 E. coli O157:H7 isolates obtained from bovine fecal samples, water trough sediment samples, free-stall bedding, and wild-bird excreta samples were characterized. Fifty genetic subtypes were observed among the isolates using XbaI and BlnI restriction endonucleases. Most restriction endonuclease digestion profiles (REDPs) were spatially and temporally clustered. However, four REDPs from multiple sources were found to be indistinguishable by pulsed-field gel electrophoresis between four pairs of farms. The geographical distance between farms which shared an indistinguishable E. coli O157:H7 REDP ranged from 9 to 50 km, and the on-farm sources sharing indistinguishable REDPs included cattle and wild bird feces and free-stall bedding. Within the study population, E. coli O157:H7 REDP subtypes were disseminated with considerable frequency among farms in close geographic proximity, and nonbovine sources may contribute to the transmission of this organism between farms.
Figures
Similar articles
-
Multiple-locus variable-nucleotide tandem repeat subtype analysis implicates European starlings as biological vectors for Escherichia coli O157:H7 in Ohio, USA.J Appl Microbiol. 2011 Oct;111(4):982-8. doi: 10.1111/j.1365-2672.2011.05102.x. Epub 2011 Aug 4. J Appl Microbiol. 2011. PMID: 21762472
-
Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover.Appl Environ Microbiol. 2004 Jan;70(1):377-84. doi: 10.1128/AEM.70.1.377-384.2004. Appl Environ Microbiol. 2004. PMID: 14711666 Free PMC article.
-
Variation in the faecal shedding of Salmonella and E. coli O157:H7 in lactating dairy cattle and examination of Salmonella genotypes using pulsed-field gel electrophoresis.Lett Appl Microbiol. 2004;38(5):366-72. doi: 10.1111/j.1472-765X.2004.01495.x. Lett Appl Microbiol. 2004. PMID: 15059205
-
Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup O157:H7.Int J Hyg Environ Health. 2001 May;203(4):347-61. doi: 10.1078/1438-4639-4410041. Int J Hyg Environ Health. 2001. PMID: 11434215 Review.
-
The control of VTEC in the animal reservoir.Int J Food Microbiol. 2001 May 21;66(1-2):71-8. doi: 10.1016/s0168-1605(00)00487-6. Int J Food Microbiol. 2001. PMID: 11407550 Review.
Cited by
-
Transmission Dynamics of Shiga Toxin-Producing Escherichia coli in New Zealand Cattle from Farm to Slaughter.Appl Environ Microbiol. 2021 May 11;87(11):e02907-20. doi: 10.1128/AEM.02907-20. Print 2021 May 11. Appl Environ Microbiol. 2021. PMID: 33771782 Free PMC article.
-
Escherichia coli O157:H7: animal reservoir and sources of human infection.Foodborne Pathog Dis. 2011 Apr;8(4):465-87. doi: 10.1089/fpd.2010.0673. Epub 2010 Nov 30. Foodborne Pathog Dis. 2011. PMID: 21117940 Free PMC article. Review.
-
Field study on evaluation of the efficacy and usability of two disinfectants for drinking water treatment at small cattle breeders and dairy cattle farms.Environ Monit Assess. 2016 Mar;188(3):151. doi: 10.1007/s10661-016-5147-0. Epub 2016 Feb 9. Environ Monit Assess. 2016. PMID: 26861741
-
Antimicrobial drug resistance against Escherichia coli and its harmful effect on animal health.Vet Med Sci. 2022 Jul;8(4):1780-1786. doi: 10.1002/vms3.825. Epub 2022 May 24. Vet Med Sci. 2022. PMID: 35608149 Free PMC article. Review.
-
E. coli O157 on Scottish cattle farms: evidence of local spread and persistence using repeat cross-sectional data.BMC Vet Res. 2014 Apr 26;10:95. doi: 10.1186/1746-6148-10-95. BMC Vet Res. 2014. PMID: 24766709 Free PMC article.
References
-
- Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 187:29-51. - PubMed
-
- Berg, J., T. McAllister, S. Bach, R. Stilborne, D. Hancock, and J. LeJeune. 2004. Escherichia coli O157:H7 excretion by commercial feedlot cattle fed either barley- or corn-based finishing diets. J. Food Prot. 67:666-671. - PubMed
-
- Brashears, M. M., M. L. Galyean, G. H. Loneragan, J. E. Mann, and K. Killinger-Mann. 2003. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 66:748-754. - PubMed
-
- Centers for Disease Control and Prevention. 2001. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7 by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, Ga.
-
- Cizek, A., P. Alexa, I. Literak, J. Hamrik, P. Novak, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435-439. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical