Identification of Sinorhizobium meliloti early symbiotic genes by use of a positive functional screen
- PMID: 16597978
- PMCID: PMC1449070
- DOI: 10.1128/AEM.72.4.2738-2748.2006
Identification of Sinorhizobium meliloti early symbiotic genes by use of a positive functional screen
Abstract
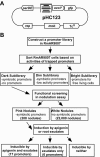
The soil bacterium Sinorhizobium meliloti establishes nitrogen-fixing symbiosis with its leguminous host plant, alfalfa, following a series of continuous signal exchanges. The complexity of the changes of alfalfa root structures during symbiosis and the amount of S. meliloti genes with unknown functions raised the possibility that more S. meliloti genes may be required for early stages of the symbiosis. A positive functional screen of the entire S. meliloti genome for symbiotic genes was carried out using a modified in vivo expression technology. A group of genes and putative genes were found to be expressed in early stages of the symbiosis, and 23 of them were alfalfa root exudate inducible. These 23 genes were further separated into two groups based on their responses to apigenin, a known nodulation (nod) gene inducer. The group of six genes not inducible by apigenin included the lsrA gene, which is essential for the symbiosis, and the dgkA gene, which is involved in the synthesis of cyclic beta-1,2-glucan required for the S. meliloti-alfalfa symbiosis. In the group of 17 apigenin-inducible genes, most have not been previously characterized in S. meliloti, and none of them belongs to the nod gene family. The identification of this large group of alfalfa root exudate-inducible S. meliloti genes suggests that the interactions in the early stages of the S. meliloti and alfalfa symbiosis could be complex and that further characterization of these genes will lead to a better understanding of the symbiosis.
Figures






Similar articles
-
How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model.Nat Rev Microbiol. 2007 Aug;5(8):619-33. doi: 10.1038/nrmicro1705. Nat Rev Microbiol. 2007. PMID: 17632573 Free PMC article. Review.
-
Contributions of Sinorhizobium meliloti Transcriptional Regulator DksA to Bacterial Growth and Efficient Symbiosis with Medicago sativa.J Bacteriol. 2016 Apr 14;198(9):1374-83. doi: 10.1128/JB.00013-16. Print 2016 May. J Bacteriol. 2016. PMID: 26883825 Free PMC article.
-
Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation.Mol Plant Microbe Interact. 2000 Jun;13(6):583-91. doi: 10.1094/MPMI.2000.13.6.583. Mol Plant Microbe Interact. 2000. PMID: 10830257
-
Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation.J Bacteriol. 2005 Jul;187(13):4562-72. doi: 10.1128/JB.187.13.4562-4572.2005. J Bacteriol. 2005. PMID: 15968067 Free PMC article.
-
Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function.Gene. 1996 Nov 7;179(1):141-6. doi: 10.1016/s0378-1119(96)00322-8. Gene. 1996. PMID: 8955640 Review.
Cited by
-
Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula.J Bacteriol. 2012 Aug;194(16):4322-31. doi: 10.1128/JB.00751-12. Epub 2012 Jun 8. J Bacteriol. 2012. PMID: 22685282 Free PMC article.
-
Gene expression analysis of the biocontrol fungus Trichoderma harzianum in the presence of tomato plants, chitin, or glucose using a high-density oligonucleotide microarray.BMC Microbiol. 2009 Oct 13;9:217. doi: 10.1186/1471-2180-9-217. BMC Microbiol. 2009. PMID: 19825185 Free PMC article.
-
Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum.Plant Physiol. 2010 Aug;153(4):1808-22. doi: 10.1104/pp.110.157800. Epub 2010 Jun 9. Plant Physiol. 2010. PMID: 20534735 Free PMC article.
-
How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model.Nat Rev Microbiol. 2007 Aug;5(8):619-33. doi: 10.1038/nrmicro1705. Nat Rev Microbiol. 2007. PMID: 17632573 Free PMC article. Review.
-
An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti.Appl Environ Microbiol. 2006 Nov;72(11):7156-67. doi: 10.1128/AEM.01397-06. Epub 2006 Sep 8. Appl Environ Microbiol. 2006. PMID: 16963549 Free PMC article.
References
-
- Becker, A., H. Berges, E. Krol, C. Bruand, S. Ruberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Kuster, C. Liebe, A. Puhler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. - PubMed
-
- Becker, A., A. Kleickmann, M. Keller, W. Arnold, and A. Puhler. 1993. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol. Gen. Genet. 241:367-379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

