Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters
- PMID: 16597980
- PMCID: PMC1449071
- DOI: 10.1128/AEM.72.4.2756-2764.2006
Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters
Abstract
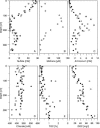
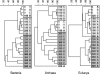
The subsurface of a tidal-flat sediment was analyzed down to 360 cm in depth by molecular and geochemical methods. A community structure analysis of all three domains of life was performed using domain-specific PCR followed by denaturing gradient gel electrophoresis analysis and sequencing of characteristic bands. The sediment column comprised horizons easily distinguishable by lithology that were deposited in intertidal and salt marsh environments. The pore water profile was characterized by a subsurface sulfate peak at a depth of about 250 cm. Methane and sulfate profiles were opposed, showing increased methane concentrations in the sulfate-free layers. The availability of organic carbon appeared to have the most pronounced effect on the bacterial community composition in deeper sediment layers. In general, the bacterial community was dominated by fermenters and syntrophic bacteria. The depth distribution of methanogenic archaea correlated with the sulfate profile and could be explained by electron donor competition with sulfate-reducing bacteria. Sequences affiliated with the typically hydrogenotrophic Methanomicrobiales were present in sulfate-free layers. Archaea belonging to the Methanosarcinales that utilize noncompetitive substrates were found along the entire anoxic-sediment column. Primers targeting the eukaryotic 18S rRNA gene revealed the presence of a subset of archaeal sequences in the deeper part of the sediment cores. The phylogenetic distance to other archaeal sequences indicates that these organisms represent a new phylogenetic group, proposed as "tidal-flat cluster 1." Eukarya were still detectable at 360 cm, even though their diversity decreased with depth. Most of the eukaryotic sequences were distantly related to those of grazers and deposit feeders.
Figures

References
-
- Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. - PubMed
-
- Böttcher, M. E., B. Hespenheide, E. Llobet-Brossa, C. Beardsley, O. Larsen, A. Schramm, A. Wieland, G. Böttcher, U.-G. Berninger, and R. Amann. 2000. The biogeochemistry, stable isotope geochemistry, and microbial community structure of a temperate intertidal mudflat: an integrated study. Continental Shelf Res. 20:1749-1769.
-
- Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb. Ecol. 36:37-50. - PubMed
-
- Chang, T. S., A. Bartholomae, E. Tilch, and B. W. Flemming. 2003. Recent development of the back-barrier tidal basin behind the island of Spiekeroog in the East Frisian Wadden Sea. Ber. Forschungszentrum Terramare 12:43-44.
-
- Coolen, M. J. L., H. Cypionka, A. M. Sass, H. Sass, and J. Overmann. 2002. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296:2407-2410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

