Chlorophenol hydroxylases encoded by plasmid pJP4 differentially contribute to chlorophenoxyacetic acid degradation
- PMID: 16597983
- PMCID: PMC1448979
- DOI: 10.1128/AEM.72.4.2783-2792.2006
Chlorophenol hydroxylases encoded by plasmid pJP4 differentially contribute to chlorophenoxyacetic acid degradation
Abstract
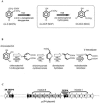
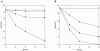
Phenoxyalkanoic compounds are used worldwide as herbicides. Cupriavidus necator JMP134(pJP4) catabolizes 2,4-dichlorophenoxyacetate (2,4-D) and 4-chloro-2-methylphenoxyacetate (MCPA), using tfd functions carried on plasmid pJP4. TfdA cleaves the ether bonds of these herbicides to produce 2,4-dichlorophenol (2,4-DCP) and 4-chloro-2-methylphenol (MCP), respectively. These intermediates can be degraded by two chlorophenol hydroxylases encoded by the tfdB(I) and tfdB(II) genes to produce the respective chlorocatechols. We studied the specific contribution of each of the TfdB enzymes to the 2,4-D/MCPA degradation pathway. To accomplish this, the tfdB(I) and tfdB(II) genes were independently inactivated, and growth on each chlorophenoxyacetate and total chlorophenol hydroxylase activity were measured for the mutant strains. The phenotype of these mutants shows that both TfdB enzymes are used for growth on 2,4-D or MCPA but that TfdB(I) contributes to a significantly higher extent than TfdB(II). Both enzymes showed similar specificity profiles, with 2,4-DCP, MCP, and 4-chlorophenol being the best substrates. An accumulation of chlorophenol was found to inhibit chlorophenoxyacetate degradation, and inactivation of the tfdB genes enhanced the toxic effect of 2,4-DCP on C. necator cells. Furthermore, increased chlorophenol production by overexpression of TfdA also had a negative effect on 2,4-D degradation by C. necator JMP134 and by a different host, Burkholderia xenovorans LB400, harboring plasmid pJP4. The results of this work indicate that codification and expression of the two tfdB genes in pJP4 are important to avoid toxic accumulations of chlorophenols during phenoxyacetic acid degradation and that a balance between chlorophenol-producing and chlorophenol-consuming reactions is necessary for growth on these compounds.
Figures


References
-
- Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology, 2nd ed. Greene Publishing Associates, New York, N.Y.
-
- Beadle, C. A., and A. R. W. Smith. 1982. The purification and properties of 2,4-dichlorophenol hydroxylase from a strain of Acinetobacter species. Eur. J. Biochem. 123:323-332. - PubMed
-
- Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

