Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment
- PMID: 16597989
- PMCID: PMC1449006
- DOI: 10.1128/AEM.72.4.2829-2836.2006
Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment
Abstract
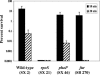
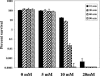
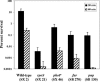
Since the stomach is a first line of defense for the host against ingested microorganisms, an ex vivo swine stomach contents (SSC) assay was developed to search for genes important for Salmonella enterica serovar Typhimurium survival in the hostile gastric environment. Initial characterization of the SSC assay (pH 3.87) using previously identified, acid-sensitive serovar Typhimurium mutants revealed a 10-fold decrease in survival for a phoP mutant following 20 min of challenge and no survival for mutants of rpoS or fur. To identify additional genes, a signature-tagged mutagenesis bank was constructed and screened in the SSC assay. Nineteen mutants were identified and individually analyzed in the SSC and acid tolerance response assays; 13 mutants exhibited a 10-fold or greater sensitivity in the SSC assay compared to the wild-type strain, but only 3 mutants displayed a 10-fold or greater decrease in survival following pH 3.0 acidic challenge. Further examination determined that the lethal effects of the SSC are pH dependent but that low pH is not the sole killing mechanism(s). Gas chromatography analysis of the SSC revealed lactic acid levels of 126 mM. Upon investigating the effects of lactic acid on serovar Typhimurium survival in a synthetic gastric fluid, not only was a concentration- and time-dependent lethal effect observed, but the phoP, rpoS, fur, and pnp genes were identified as involved in protection against lactic acid exposure. These studies indicate a role in gastric survival for several serovar Typhimurium genes and imply that the stomach environment is defined by more than low pH.
Figures


Similar articles
-
The lactic acid-induced acid tolerance response in Salmonella enterica serovar Typhimurium induces sensitivity to hydrogen peroxide.Appl Environ Microbiol. 2006 Aug;72(8):5623-5. doi: 10.1128/AEM.00538-06. Appl Environ Microbiol. 2006. PMID: 16885318 Free PMC article.
-
Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation.Infect Immun. 2001 Oct;69(10):6463-74. doi: 10.1128/IAI.69.10.6463-6474.2001. Infect Immun. 2001. PMID: 11553591 Free PMC article.
-
Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose.Appl Environ Microbiol. 2006 Jul;72(7):5002-12. doi: 10.1128/AEM.02868-05. Appl Environ Microbiol. 2006. PMID: 16820499 Free PMC article.
-
Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection.FEMS Microbiol Rev. 2005 Nov;29(5):1021-40. doi: 10.1016/j.femsre.2005.03.005. Epub 2005 Jun 29. FEMS Microbiol Rev. 2005. PMID: 16023758 Review.
-
Differential gene expression of pathogens inside infected hosts.Curr Opin Microbiol. 2006 Apr;9(2):138-42. doi: 10.1016/j.mib.2006.01.003. Epub 2006 Feb 3. Curr Opin Microbiol. 2006. PMID: 16459132 Review.
Cited by
-
Salmonella pathogenicity and host adaptation in chicken-associated serovars.Microbiol Mol Biol Rev. 2013 Dec;77(4):582-607. doi: 10.1128/MMBR.00015-13. Microbiol Mol Biol Rev. 2013. PMID: 24296573 Free PMC article. Review.
-
Dissecting the Acid Stress Response of Rhizobium tropici CIAT 899.Front Microbiol. 2018 Apr 30;9:846. doi: 10.3389/fmicb.2018.00846. eCollection 2018. Front Microbiol. 2018. PMID: 29760688 Free PMC article.
-
The bacterial translation stress response.FEMS Microbiol Rev. 2014 Nov;38(6):1172-201. doi: 10.1111/1574-6976.12083. Epub 2014 Sep 26. FEMS Microbiol Rev. 2014. PMID: 25135187 Free PMC article. Review.
-
Clearance of Streptococcus suis in Stomach Contents of Differently Fed Growing Pigs.Pathogens. 2016 Aug 6;5(3):56. doi: 10.3390/pathogens5030056. Pathogens. 2016. PMID: 27509526 Free PMC article.
-
Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit.Appl Environ Microbiol. 2008 Mar;74(5):1583-97. doi: 10.1128/AEM.01507-07. Epub 2008 Jan 11. Appl Environ Microbiol. 2008. PMID: 18192414 Free PMC article.
References
-
- Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. - PubMed
-
- Baik, H. S., S. Bearson, S. Dunbar, and J. W. Foster. 1996. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology 142:3195-3200. - PubMed
-
- Barber, D. A., P. B. Bahnson, R. Isaacson, C. J. Jones, and R. M. Weigel. 2002. Distribution of Salmonella in swine production ecosystems. J. Food Prot. 65:1861-1868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

