Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities
- PMID: 16598010
- PMCID: PMC1449077
- DOI: 10.1128/AEM.72.4.3016-3020.2006
Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities
Abstract
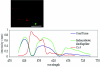
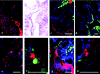
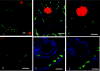
Microbial communities play important roles in the functioning of coral reef communities. However, extensive autofluorescence of coral tissues and endosymbionts limits the application of standard fluorescence in situ hybridization (FISH) techniques for the identification of the coral-associated bacterial communities. This study overcomes these limitations by combining FISH and spectral imaging.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

