Identification and characterization of CRT10 as a novel regulator of Saccharomyces cerevisiae ribonucleotide reductase genes
- PMID: 16600900
- PMCID: PMC1447646
- DOI: 10.1093/nar/gkl100
Identification and characterization of CRT10 as a novel regulator of Saccharomyces cerevisiae ribonucleotide reductase genes
Abstract
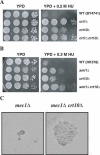
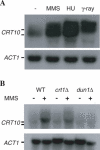
The CRT10 gene was identified through screening of the Saccharomyces cerevisiae deletion library for hydroxyurea (HU) resistance. CRT10 encodes a putative 957 amino acid, 110 kDa protein with a leucine repeat and a WD40 repeat near the N-terminus. Deletion of CRT10 resulted in an enhanced resistance to HU reminiscent of the inactivation of two other ribonucleotide reductase (Rnr) suppressors, CRT1 and SML1, which regulate Rnr activity at transcriptional and translational levels, respectively. Epistatic analysis indicates that CRT10 belongs to the CRT1 pathway but not the SML1 pathway. Indeed, deletion of CRT10 enhanced the survival of the mec1 null mutant and increased basal level and DNA damage-induced expression of RNR2 and RNR3, suggesting that Crt10 regulates RNR genes at the transcriptional level. Furthermore, the dun1 mutation is epistatic to crt10 with respect to both HU sensitivity and RNR gene expression. Interestingly, the expression of CRT10 itself is induced by DNA damaging agents and this induction requires DUN1, suggesting that CRT10 plays a role in cellular response to DNA damage and replication blocks. The CRT10 function appears to be achieved by positive regulation of the CRT1 transcript level, indicating that CRT10 is a component of the regulatory circuit.
Figures




Similar articles
-
Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools.PLoS Genet. 2011 May;7(5):e1002061. doi: 10.1371/journal.pgen.1002061. Epub 2011 May 5. PLoS Genet. 2011. PMID: 21573136 Free PMC article.
-
Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length.J Cell Sci. 2006 Dec 15;119(Pt 24):5178-92. doi: 10.1242/jcs.03221. J Cell Sci. 2006. PMID: 17158920
-
Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae.Mol Cell Biol. 1997 Oct;17(10):6105-13. doi: 10.1128/MCB.17.10.6105. Mol Cell Biol. 1997. PMID: 9315670 Free PMC article.
-
DNA damage and cell cycle regulation of ribonucleotide reductase.Bioessays. 1993 May;15(5):333-9. doi: 10.1002/bies.950150507. Bioessays. 1993. PMID: 8343143 Review.
-
Rnr1's role in telomere elongation cannot be replaced by Rnr3: a role beyond dNTPs?Curr Genet. 2018 Jun;64(3):547-550. doi: 10.1007/s00294-017-0779-3. Epub 2017 Nov 8. Curr Genet. 2018. PMID: 29119271 Review.
Cited by
-
Mms1 binds to G-rich regions in Saccharomyces cerevisiae and influences replication and genome stability.Nucleic Acids Res. 2017 Jul 27;45(13):7796-7806. doi: 10.1093/nar/gkx467. Nucleic Acids Res. 2017. PMID: 28535251 Free PMC article.
-
The mutation of a novel Saccharomyces cerevisiae SRL4 gene rescues the lethality of rad53 and lcd1 mutations by modulating dNTP levels.J Microbiol. 2008 Feb;46(1):75-80. doi: 10.1007/s12275-008-0013-6. J Microbiol. 2008. PMID: 18337697
-
The HMGB Protein KlIxr1, a DNA Binding Regulator of Kluyveromyces lactis Gene Expression Involved in Oxidative Metabolism, Growth, and dNTP Synthesis.Biomolecules. 2021 Sep 21;11(9):1392. doi: 10.3390/biom11091392. Biomolecules. 2021. PMID: 34572607 Free PMC article.
-
Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis.Plant Physiol. 2009 Sep;151(1):461-71. doi: 10.1104/pp.109.140053. Epub 2009 Jul 1. Plant Physiol. 2009. PMID: 19571309 Free PMC article.
-
Physical interactions between specifically regulated subpopulations of the MCM and RNR complexes prevent genetic instability.PLoS Genet. 2024 May 22;20(5):e1011148. doi: 10.1371/journal.pgen.1011148. eCollection 2024 May. PLoS Genet. 2024. PMID: 38776358 Free PMC article.
References
-
- Jordan A., Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 1998;67:71–98. - PubMed
-
- Eklund H., Uhlin U., Farnegardh M., Logan D.T., Nordlund P. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 2001;77:177–268. - PubMed
-
- Fontecave M., Nordlund P., Eklund H., Reichard P. The redox centers of ribonucleotide reductase of Escherichia coli. Adv. Enzymol. Relat. Areas Mol. Biol. 1992;65:147–183. - PubMed
-
- Elledge S.J., Davis R.W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

