Thermodynamics of unfolding of an integral membrane protein in mixed micelles
- PMID: 16600971
- PMCID: PMC2242483
- DOI: 10.1110/ps.052031306
Thermodynamics of unfolding of an integral membrane protein in mixed micelles
Abstract
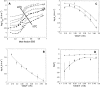
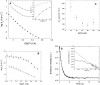
Quantitative studies of membrane protein folding and unfolding can be difficult because of difficulties with efficient refolding as well as a pronounced propensity to aggregate. However, mixed micelles, consisting of the anionic detergent sodium dodecyl sulfate and the nonionic detergent dodecyl maltoside facilitate reversible and quantitative unfolding and refolding. The 4-transmembrane helix protein DsbB from the inner membrane of Escherichia coli unfolds in mixed micelles according to a three-state mechanism involving an unfolding intermediate I. The temperature dependence of the kinetics of this reaction between 15 degrees and 45 degrees C supports that unfolding from I to the denatured state D is accompanied by a significant decrease in heat capacity. For water-soluble proteins, the heat capacity increases upon unfolding, and this is generally interpreted as the increased binding of water to the protein as it unfolds, exposing more surface area. The decrease in DsbB's heat capacity upon unfolding is confirmed by independent thermal scans. The decrease in heat capacity is not an artifact of the use of mixed micelles, since the water soluble protein S6 shows conventional heat-capacity changes in detergent. We speculate that it reflects the binding of SDS to parts of DsbB that are solvent-exposed in the native DM-bound state. This implies that the periplasmic loops of DsbB are relatively unstructured. This anomalous thermodynamic behavior has not been observed for beta-barrel membrane proteins, probably because they do not bind SDS so extensively. Thus the thermodynamic behavior of membrane proteins appears to be intimately connected to their detergent-binding properties.
Figures

References
-
- Aniansson E.G. and Wall S.N. 1974. On the kinetics of stepwise micelle association J. Phys. Chem. 78: 1024–1030.
-
- Brouillette C.G., Muccio D.D., Finney T.K. 1987. pH dependence of bacteriorhodopsin thermal unfolding Biochemistry 26: 7431–7438. - PubMed
-
- Creighton T.E. In Proteins. Structures and molecular properties . 1993. 2d ed. W.H. Freeman & Co, New York.
-
- Faham S., Yang D., Bare E., Yohannan S., Whitelegge J.P., Bowie J.U. 2004. Side-chain contributions to membrane protein structure and stability J. Mol. Biol. 335: 297–305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

