Identification of a pH sensor in the furin propeptide that regulates enzyme activation
- PMID: 16601116
- PMCID: PMC4293020
- DOI: 10.1074/jbc.M600760200
Identification of a pH sensor in the furin propeptide that regulates enzyme activation
Abstract
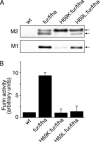
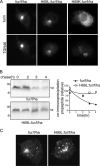
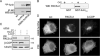
The folding and activation of furin occur through two pH- and compartment-specific autoproteolytic steps. In the endoplasmic reticulum (ER), profurin folds under the guidance of its prodomain and undergoes an autoproteolytic excision at the consensus furin site Arg-Thr-Lys-Arg107/ generating an enzymatically masked furin-propeptide complex competent for transport to late secretory compartments. In the mildly acidic environment of the trans-Golgi network/endosomal system, the bound propeptide is cleaved at the internal site 69HRGVTKR75/, unmasking active furin capable of cleaving substrates in trans. Here, by using cellular, biochemical, and modeling studies, we demonstrate that the conserved His69 is a pH sensor that regulates the compartment-specific cleavages of the propeptide. In the ER, unprotonated His69 stabilizes a solvent-accessible hydrophobic pocket necessary for autoproteolytic excision at Arg107. Profurin molecules unable to form the hydrophobic pocket, and hence, the furin-propeptide complex, are restricted to the ER by a PACS-2- and COPI-dependent mechanism. Once exposed to the acidic pH of the late secretory pathway, protonated His69 disrupts the hydrophobic pocket, resulting in exposure and cleavage of the internal cleavage site at Arg75 to unmask the enzyme. Together, our data explain the pH-regulated activation of furin and how this His-dependent regulatory mechanism is a model for other proteins.
Figures


Similar articles
-
The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation.J Biol Chem. 2002 Apr 12;277(15):12879-90. doi: 10.1074/jbc.M108740200. Epub 2002 Jan 17. J Biol Chem. 2002. PMID: 11799113 Free PMC article.
-
Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage.EMBO J. 1997 Apr 1;16(7):1508-18. doi: 10.1093/emboj/16.7.1508. EMBO J. 1997. PMID: 9130696 Free PMC article.
-
Endoproteolytic cleavage of its propeptide is a prerequisite for efficient transport of furin out of the endoplasmic reticulum.J Biol Chem. 1995 Feb 10;270(6):2695-702. doi: 10.1074/jbc.270.6.2695. J Biol Chem. 1995. PMID: 7852339
-
[Furin and its biological role].Ukr Biokhim Zh (1999). 2007 Nov-Dec;79(6):5-18. Ukr Biokhim Zh (1999). 2007. PMID: 18712106 Review. Russian.
-
Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins.Biochem J. 1997 Nov 1;327 ( Pt 3)(Pt 3):625-35. doi: 10.1042/bj3270625. Biochem J. 1997. PMID: 9599222 Free PMC article. Review.
Cited by
-
Cysteine Proteases: Modes of Activation and Future Prospects as Pharmacological Targets.Front Pharmacol. 2016 Apr 25;7:107. doi: 10.3389/fphar.2016.00107. eCollection 2016. Front Pharmacol. 2016. PMID: 27199750 Free PMC article. Review.
-
Autocatalytic activation of the furin zymogen requires removal of the emerging enzyme's N-terminus from the active site.PLoS One. 2009;4(4):e5031. doi: 10.1371/journal.pone.0005031. Epub 2009 Apr 7. PLoS One. 2009. PMID: 19352504 Free PMC article.
-
The controversial effect of smoking and nicotine in SARS-CoV-2 infection.Allergy Asthma Clin Immunol. 2023 Jun 1;19(1):49. doi: 10.1186/s13223-023-00797-0. Allergy Asthma Clin Immunol. 2023. PMID: 37264452 Free PMC article. Review.
-
Structurally Modified Bioactive Peptide Inhibits SARS-CoV-2 Lentiviral Particles Expression.Pharmaceutics. 2022 Sep 26;14(10):2045. doi: 10.3390/pharmaceutics14102045. Pharmaceutics. 2022. PMID: 36297481 Free PMC article.
-
The Path to Therapeutic Furin Inhibitors: From Yeast Pheromones to SARS-CoV-2.Int J Mol Sci. 2022 Mar 22;23(7):3435. doi: 10.3390/ijms23073435. Int J Mol Sci. 2022. PMID: 35408793 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

