Assessing IRES activity in the HIF-1alpha and other cellular 5' UTRs
- PMID: 16601206
- PMCID: PMC1464860
- DOI: 10.1261/rna.2320506
Assessing IRES activity in the HIF-1alpha and other cellular 5' UTRs
Abstract
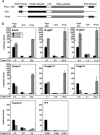
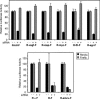
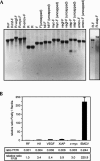
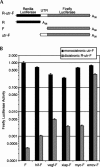
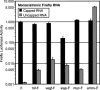
Dicistronic reporter plasmids, such as the dual luciferase-containing pR-F plasmid, have been widely used to assay cellular and viral 5' untranslated regions (UTRs) for IRES activity. We found that the pR-F dicistronic reporter containing the 5' UTRs from HIF-1alpha, VEGF, c-myc, XIAP, VEGFR-1, or Egr-1 UTRs all produce the downstream luciferase predominantly as a result of cryptic promoter activity that is activated by the SV40 enhancer elements in the plasmid. RNA transfection experiments using dicistronic or uncapped RNAs, which avoid the complication of cryptic promoter activity, indicate that the HIF-1alpha, VEGF, c-myc, and XIAP UTRs do have some IRES activity, although the activity was much less than that of the viral EMCV IRES. The translation of transfected monocistronic RNAs containing these cellular UTRs was greatly enhanced by the presence of a 5' cap, raising questions as to the strength or mechanism of IRES-mediated translation in these assays.
Figures


References
-
- Akiri G., Nahari D., Finkelstein Y., Le S.Y., Elroy-Stein O., Levi B.Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. - PubMed
-
- Boshart M., Weber F., Jahn G., Dorsch-Hasler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. - PubMed
-
- Byrd M.P., Zamora M., Lloyd R.E. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J. Biol. Chem. 2005;280:18610–18622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
