Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2
- PMID: 16601676
- PMCID: PMC1456944
- DOI: 10.1038/sj.emboj.7601079
Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2
Abstract
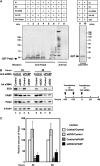
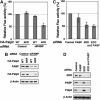
The poly(A)-binding protein (PABP) is a unique translation initiation factor in that it binds to the mRNA 3' poly(A) tail and stimulates recruitment of the ribosome to the mRNA at the 5' end. PABP activity is tightly controlled by the PABP-interacting protein 2 (Paip2), which inhibits translation by displacing PABP from the mRNA. Here, we describe a close interplay between PABP and Paip2 protein levels in the cell. We demonstrate a mechanism for this co-regulation that involves an E3 ubiquitin ligase, EDD, which targets Paip2 for degradation. PABP depletion by RNA interference (RNAi) causes co-depletion of Paip2 protein without affecting Paip2 mRNA levels. Upon PABP knockdown, Paip2 interacts with EDD, which leads to Paip2 ubiquitination. Supporting a critical role for EDD in Paip2 degradation, knockdown of EDD expression by siRNA leads to an increase in Paip2 protein stability. Thus, we demonstrate that the turnover of Paip2 in the cell is mediated by EDD and is regulated by PABP. This mechanism serves as a homeostatic feedback to control the activity of PABP in cells.
Figures






References
-
- Adams J (2001) Proteasome inhibition in cancer: development of PS-341. Semin Oncol 28: 613–619 - PubMed
-
- Bonifacino JS, Cosson P, Klausner RD (1990) Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63: 503–513 - PubMed
-
- Callaghan MJ, Russell AJ, Woollatt E, Sutherland GR, Sutherland RL, Watts CK (1998) Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17: 3479–3491 - PubMed
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

