Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses
- PMID: 16601681
- PMCID: PMC1440839
- DOI: 10.1038/sj.emboj.7601073
Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses
Abstract
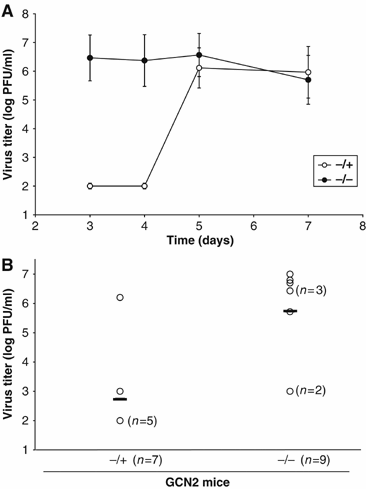
In mammals, four different protein kinases, heme-regulated inhibitor, double-stranded RNA-dependent protein kinase (PKR), general control non-derepressible-2 (GCN2) and PKR-like endoplasmic reticulum kinase, regulate protein synthesis in response to environmental stresses by phosphorylating the alpha-subunit of the initiation factor 2 (eIF2alpha). We now report that mammalian GCN2 is specifically activated in vitro upon binding of two nonadjacent regions of the Sindbis virus (SV) genomic RNA to its histidyl-tRNA synthetase-related domain. Moreover, endogenous GCN2 is activated in cells upon SV infection. Strikingly, fibroblasts derived from GCN2-/- mice possess an increased permissiveness to SV or vesicular stomatitis virus infection. We further show that mice lacking GCN2 are extremely susceptible to intranasal SV infection, demonstrating high virus titers in the brain compared to similarly infected control animals. The overexpression of wild-type GCN2, but not the catalytically inactive GCN2-K618R variant, in NIH 3T3 cells impaired the replication of a number of RNA viruses. We determined that GCN2 inhibits SV replication by blocking early viral translation of genomic SV RNA. These findings point to a hitherto unrecognized role of GCN2 as an early mediator in the cellular response to RNA viruses.
Figures



References
-
- Abraham N, Stojdl DF, Duncan PI, Methot N, Ishii T, Dube M, Vanderhyden BC, Atkins HL, Gray DA, McBurney MW, Koromilas AE, Brown EG, Sonenberg N, Bell JC (1999) Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem 274: 5953–5962 - PubMed
-
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN (2000) Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13: 129–141 - PubMed
-
- Berlanga JJ, Herrero S, de Haro C (1998) Characterization of the hemin-sensitive eukaryotic initiation factor 2α kinase from mouse nonerythroid cells. J Biol Chem 273: 32340–32346 - PubMed
-
- Berlanga JJ, Santoyo J, de Haro C (1999) Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur J Biochem 265: 754–762 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

