Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks
- PMID: 16601689
- PMCID: PMC1440835
- DOI: 10.1038/sj.emboj.7601063
Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks
Abstract
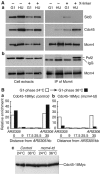
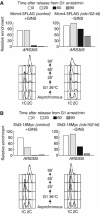
The Cdc45 protein is crucial for the initiation of chromosome replication in eukaryotic cells, as it allows the activation of prereplication complexes (pre-RCs) that contain the MCM helicase. This causes the unwinding of origins and the establishment of DNA replication forks. The incorporation of Cdc45 at nascent forks is a highly regulated and poorly understood process that requires, in budding yeast, the Sld3 protein and the GINS complex. Previous studies suggested that Sld3 is also important for the progression of DNA replication forks after the initiation step, as are Cdc45 and GINS. In contrast, we show here that Sld3 does not move with DNA replication forks and only associates with MCM in an unstable manner before initiation. After the establishment of DNA replication forks from early origins, Sld3 is no longer essential for the completion of chromosome replication. Unlike Sld3, GINS is not required for the initial recruitment of Cdc45 to origins and instead is necessary for stable engagement of Cdc45 with the nascent replisome. Like Cdc45, GINS then associates stably with MCM during S-phase.
Figures






References
-
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM complexes and Cdc45p during S phase. Cell 91: 59–69 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

