Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder
- PMID: 16602696
- PMCID: PMC2533134
- DOI: 10.1021/pr060049p
Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder
Abstract
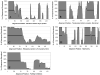
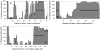
Regions of conserved disorder prediction (CDP) were found in protein domains from all available InterPro member databases, although with varying frequency. These CDP regions were found in proteins from all kingdoms of life, including viruses. However, eukaryotes had 1 order of magnitude more proteins containing long disordered regions than did archaea and bacteria. Sequence conservation in CDP regions varied, but was on average slightly lower than in regions of conserved order. In some cases, disordered regions evolve faster than ordered regions, in others they evolve slower, and in the rest they evolve at roughly the same rate. A variety of functions were found to be associated with domains containing conserved disorder. The most common were DNA/RNA binding, and protein binding. Many ribosomal proteins also were found to contain conserved disordered regions. Other functions identified included membrane translocation and amino acid storage for germination. Due to limitations of current knowledge as well as the methodology used for this work, it was not determined whether these functions were directly associated with the predicted disordered region. However, the functions associated with conserved disorder in this work are in agreement with the functions found in other studies to correlate to disordered regions. We have established that intrinsic disorder may be more common in bacterial and archaeal proteins than previously thought, but this disorder is likely to be used for different purposes than in eukaryotic proteins, as well as occurring in shorter stretches of protein. Regions of predicted disorder were found to be conserved within a large number of protein families and domains. Although many think of such conserved domains as being ordered, in fact a significant number of them contain regions of disorder that are likely to be crucial to their functions.
Figures
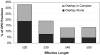




References
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. - PubMed
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. - PubMed
-
- Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. - PubMed
-
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

