Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence
- PMID: 16603661
- PMCID: PMC1475451
- DOI: 10.1104/pp.106.079293
Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence
Abstract
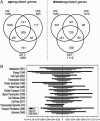
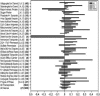
A comparative transcriptome analysis for successive stages of Arabidopsis (Arabidopsis thaliana) developmental leaf senescence (NS), darkening-induced senescence of individual leaves attached to the plant (DIS), and senescence in dark-incubated detached leaves (DET) revealed many novel senescence-associated genes with distinct expression profiles. The three senescence processes share a high number of regulated genes, although the overall number of regulated genes during DIS and DET is about 2 times lower than during NS. Consequently, the number of NS-specific genes is much higher than the number of DIS- or DET-specific genes. The expression profiles of transporters (TPs), receptor-like kinases, autophagy genes, and hormone pathways were analyzed in detail. The Arabidopsis TPs and other integral membrane proteins were systematically reclassified based on the Transporter Classification system. Coordinate activation or inactivation of several genes is observed in some TP families in all three or only in individual senescence types, indicating differences in the genetic programs for remobilization of catabolites. Characteristic senescence type-specific differences were also apparent in the expression profiles of (putative) signaling kinases. For eight hormones, the expression of biosynthesis, metabolism, signaling, and (partially) response genes was investigated. In most pathways, novel senescence-associated genes were identified. The expression profiles of hormone homeostasis and signaling genes reveal additional players in the senescence regulatory network.
Figures





Similar articles
-
A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves.Plant J. 2009 Feb;57(4):690-705. doi: 10.1111/j.1365-313X.2008.03722.x. Epub 2008 Oct 16. Plant J. 2009. PMID: 18980641
-
AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana.Int J Mol Sci. 2021 May 5;22(9):4885. doi: 10.3390/ijms22094885. Int J Mol Sci. 2021. PMID: 34063046 Free PMC article.
-
Dissecting the Metabolic Role of Mitochondria during Developmental Leaf Senescence.Plant Physiol. 2016 Dec;172(4):2132-2153. doi: 10.1104/pp.16.01463. Epub 2016 Oct 15. Plant Physiol. 2016. PMID: 27744300 Free PMC article.
-
Transcription factors regulating leaf senescence in Arabidopsis thaliana.Plant Biol (Stuttg). 2008 Sep;10 Suppl 1:63-75. doi: 10.1111/j.1438-8677.2008.00088.x. Plant Biol (Stuttg). 2008. PMID: 18721312 Review.
-
A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations.J Exp Bot. 2018 Feb 12;69(4):769-786. doi: 10.1093/jxb/erx246. J Exp Bot. 2018. PMID: 28992225 Review.
Cited by
-
Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease.Plant Physiol. 2012 Nov;160(3):1432-52. doi: 10.1104/pp.112.204362. Epub 2012 Sep 6. Plant Physiol. 2012. PMID: 22961132 Free PMC article.
-
Transport proteins regulate the flux of metabolites and cofactors across the membrane of plant peroxisomes.Front Plant Sci. 2012 Jan 16;3:3. doi: 10.3389/fpls.2012.00003. eCollection 2012. Front Plant Sci. 2012. PMID: 22645564 Free PMC article.
-
The AtMINPP Gene, Encoding a Multiple Inositol Polyphosphate Phosphatase, Coordinates a Novel Crosstalk between Phytic Acid Metabolism and Ethylene Signal Transduction in Leaf Senescence.Int J Mol Sci. 2024 Aug 17;25(16):8969. doi: 10.3390/ijms25168969. Int J Mol Sci. 2024. PMID: 39201658 Free PMC article.
-
Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity.J Exp Bot. 2010 Mar;61(5):1419-30. doi: 10.1093/jxb/erq010. Epub 2010 Feb 17. J Exp Bot. 2010. PMID: 20164142 Free PMC article.
-
Signal Transduction in Leaf Senescence: Progress and Perspective.Plants (Basel). 2019 Oct 10;8(10):405. doi: 10.3390/plants8100405. Plants (Basel). 2019. PMID: 31658600 Free PMC article. Review.
References
-
- Addicott FT, Lynch RS, Carns HR (1955) Auxin gradient theory of abscission regulation. Science 121: 644–645 - PubMed
-
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41: 859–874 - PubMed
-
- Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al (2004) Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16: 1925–1937 - PMC - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

