Functional analysis of a mammalian odorant receptor subfamily
- PMID: 16606354
- PMCID: PMC4096696
- DOI: 10.1111/j.1471-4159.2006.03859.x
Functional analysis of a mammalian odorant receptor subfamily
Abstract
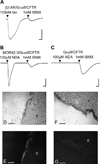
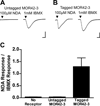
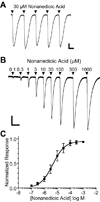
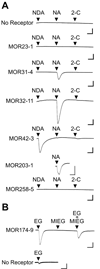
Phylogenetic analysis groups mammalian odorant receptors into two broad classes and numerous subfamilies. These subfamilies are proposed to reflect functional organization. Testing this idea requires an assay allowing detailed functional characterization of odorant receptors. Here we show that a variety of Class I and Class II mouse odorant receptors can be functionally expressed in Xenopus laevis oocytes. Receptor constructs included the N-terminal 20 residues of human rhodopsin and were co-expressed with Galphaolf and the cystic fibrosis transmembrane regulator to allow electrophysiological measurement of receptor responses. For most mouse odorant receptors tested, these conditions were sufficient for functional expression. Co-expression of accessory proteins was required to allow functional surface expression of some mouse odorant receptors. We used this assay to examine the receptive ranges of all members of the mouse odorant receptor 42 (MOR42) subfamily. MOR42-1 responded to dicarboxylic acids, preferring a 10-12 carbon chain length. MOR42-2 responded to monocarboxylic acids (7-10 carbons). MOR42-3 responded to dicarboxylic acids (8-10 carbons) and monocarboxylic acids (10-12 carbons). Thus, the receptive range of each receptor was unique. However, overlap between the individual receptive ranges suggests that the members of this subfamily form one contiguous subfamily receptive range, suggesting that odorant receptor subfamilies do constitute functional units.
Figures


Similar articles
-
The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues.J Biol Chem. 2007 Jan 12;282(2):1216-24. doi: 10.1074/jbc.M609355200. Epub 2006 Nov 17. J Biol Chem. 2007. PMID: 17114180
-
Mammalian odorant receptor tuning breadth persists across distinct odorant panels.PLoS One. 2017 Sep 25;12(9):e0185329. doi: 10.1371/journal.pone.0185329. eCollection 2017. PLoS One. 2017. PMID: 28945824 Free PMC article.
-
Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids.J Neurochem. 2009 Apr;109(1):193-202. doi: 10.1111/j.1471-4159.2009.05925.x. Epub 2009 Feb 23. J Neurochem. 2009. PMID: 19166503 Free PMC article.
-
Early events in olfaction: diversity and spatial patterns of odorant receptors.Recept Channels. 1993;1(4):259-66. Recept Channels. 1993. PMID: 8081724 Review.
-
Molecular biology of smell: expression of the multigene family encoding putative odorant receptors.Cold Spring Harb Symp Quant Biol. 1992;57:505-16. doi: 10.1101/sqb.1992.057.01.056. Cold Spring Harb Symp Quant Biol. 1992. PMID: 1339687 Review. No abstract available.
Cited by
-
How does your kidney smell? Emerging roles for olfactory receptors in renal function.Pediatr Nephrol. 2016 May;31(5):715-23. doi: 10.1007/s00467-015-3181-8. Epub 2015 Aug 12. Pediatr Nephrol. 2016. PMID: 26264790 Free PMC article. Review.
-
Predatory mite attraction to herbivore-induced plant odors is not a consequence of attraction to individual herbivore-induced plant volatiles.J Chem Ecol. 2008 Jun;34(6):791-803. doi: 10.1007/s10886-008-9492-5. Epub 2008 Jun 3. J Chem Ecol. 2008. PMID: 18521678 Free PMC article.
-
Structure-activity relationships on the odor detectability of homologous carboxylic acids by humans.Exp Brain Res. 2010 Nov;207(1-2):75-84. doi: 10.1007/s00221-010-2430-0. Epub 2010 Oct 8. Exp Brain Res. 2010. PMID: 20931179 Free PMC article.
-
Functional analysis of human olfactory receptors with a high basal activity using LNCaP cell line.PLoS One. 2022 Apr 21;17(4):e0267356. doi: 10.1371/journal.pone.0267356. eCollection 2022. PLoS One. 2022. PMID: 35446888 Free PMC article.
-
Dietary Suberic Acid Protects Against UVB-Induced Skin Photoaging in Hairless Mice.Nutrients. 2019 Dec 4;11(12):2948. doi: 10.3390/nu11122948. Nutrients. 2019. PMID: 31817085 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

