Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors
- PMID: 16606439
- PMCID: PMC1557996
- DOI: 10.1186/gb-2006-7-4-r28
Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors
Abstract
Background: Estrogen plays a central role in breast cancer pathogenesis. Although many studies have characterized the estrogen regulation of genes using in vitro cell culture models by global mRNA expression profiling, it is not clear whether these genes are similarly regulated in vivo or how they might be coordinately expressed in primary human tumors.
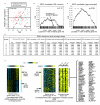
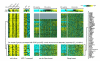
Results: We generated DNA microarray-based gene expression profiles from three estrogen receptor alpha (ERalpha)-positive breast cancer cell lines stimulated by 17beta-estradiol (E2) in vitro over a time course, as well as from MCF-7 cells grown as xenografts in ovariectomized athymic nude mice with E2 supplementation and after its withdrawal. When the patterns of genes regulated by E2 in vitro were compared to those obtained from xenografts, we found a remarkable overlap (over 40%) of genes regulated by E2 in both contexts. These patterns were compared to those obtained from published clinical data sets. We show that, as a group, E2-regulated genes from our preclinical models were co-expressed with ERalpha in a panel of ERalpha+ breast tumor mRNA profiles, when corrections were made for patient age, as well as with progesterone receptor. Furthermore, the E2-regulated genes were significantly enriched for transcriptional targets of the myc oncogene and were found to be coordinately expressed with Myc in human tumors.
Conclusion: Our results provide significant validation of a widely used in vitro model of estrogen signaling as being pathologically relevant to breast cancers in vivo.
Figures


References
-
- Elledge RM, Fuqua SA. Estrogen and progesterone receptors. In: Harris J, Lippman ME, Morrow M, Osborne C, editor. Diseases of the Breast. Philadelphia: Lippincott, Williams and Wilkins; 2000. pp. 471–488.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

