Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1
- PMID: 16606612
- PMCID: PMC4196876
- DOI: 10.1074/jbc.M600807200
Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1
Abstract
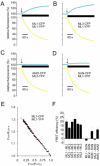
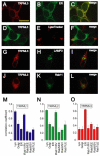
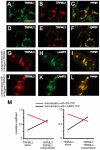
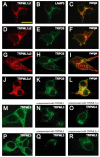
Mucolipidosis type IV is an autosomal recessive lysosomal storage disorder characterized by severe neurodegeneration, achlorhydria, and visual impairments such as corneal opacity and strabismus. The disease arises due to mutations in a group 2 transient receptor potential (TRP)-related cation channel, TRPML1. Mammals encode two additional TRPML proteins named TRPML2 and TRPML3. Information regarding the propensity of these proteins to multimerize, their subcellular distribution and mechanisms that regulate their trafficking are limited. Here we demonstrate that TRPMLs interact to form homo- and heteromultimers. Moreover, the presence of either TRPML1 or TRPML2 specifically influences the spatial distribution of TRPML3. TRPML1 and TRPML2 homomultimers are lysosomal proteins, whereas TRPML3 homomultimers are in the endoplasmic reticulum. However, TRPML3 localizes to lysosomes when coexpressed with either TRPML1 or TRPML2 and is comparably mislocalized when lysosomal targeting of TRPML1 and TRPML2 is disrupted. Conversely, TRPML3 does not cause retention of TRPML1 or TRPML2 in the endoplasmic reticulum. These data demonstrate that there is a hierarchy controlling the subcellular distributions of the TRPMLs such that TRPML1 and TRPML2 dictate the localization of TRPML3 and not vice versa.
Figures


References
-
- Bach G. Mol. Genet. Metab. 2001;73:197–203. - PubMed
-
- Bach G. Pfluegers. Arch. 2005;451:313–317. - PubMed
-
- Altarescu G, Sun M, Moore DF, Smith JA, Wiggs EA, Solomon BI, Patronas NJ, Frei KP, Gupta S, Kaneski CR, Quarrell OW, Slaugenhaupt SA, Goldin E, Schiffmann R. Neurology. 2002;59:306–313. - PubMed
-
- Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, Kiselyov K. J. Biol. Chem. 2006;281:7294–7301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

