Conservation and functional significance of gene topology in the genome of Caenorhabditis elegans
- PMID: 16606698
- PMCID: PMC1457050
- DOI: 10.1101/gr.4515306
Conservation and functional significance of gene topology in the genome of Caenorhabditis elegans
Abstract
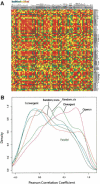
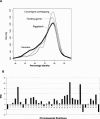
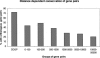
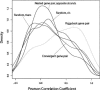
We have systematically examined the correlation between transcriptional expression pattern and the physical layout of gene pairs in the genome of Caenorhabditis elegans using a public tissue-specific SAGE library data set. We find a strong positive correlation in the expression patterns of neighboring gene pairs that are close together and transcribed in the same direction as well as for neighboring pairs that are located on opposing strands and transcribed in divergent directions. Coupling between members of nonoverlapping neighboring gene pairs is independent of operons and decreases to background levels as the distance increases beyond 10 kb. These findings suggest the existence of regional transcriptional domains in the C. elegans genome. In contrast, genes that are on opposing strands and transcribed in convergent directions are less transcriptionally coupled than the genome-wide background, suggesting a mutual inhibition mechanism. We have also examined the conservation and functional consequences of extreme cases of topological entanglement in the C. elegans genome, in which two or more genes physically overlap in their UTRs or coding regions. We have found that overlapping gene pairs are more conserved and are enriched in essential genes and genes that cause various defined phenotypes revealed by RNAi trials. SAGE analysis indicates that genes that are on the same strand, physically overlap, and transcript at the same directions are very highly correlated in gene expression, while overlapping gene pairs in which one member of the pair resides within an intron of the other are weakly, if at all, coupled, similar to convergent overlapping genes.
Figures



References
-
- Blumenthal T. Operons in eukaryotes. Brief Funct. Genomic Proteomic. 2004;3:199–211. - PubMed
-
- Blumenthal T., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Chiu W.L., Duke K., Kiraly M., Duke K., Kiraly M., Kiraly M., et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. - PubMed
-
- C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
-
- Celniker S.E., Rubin G.M., Rubin G.M. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 2003;4:89–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
